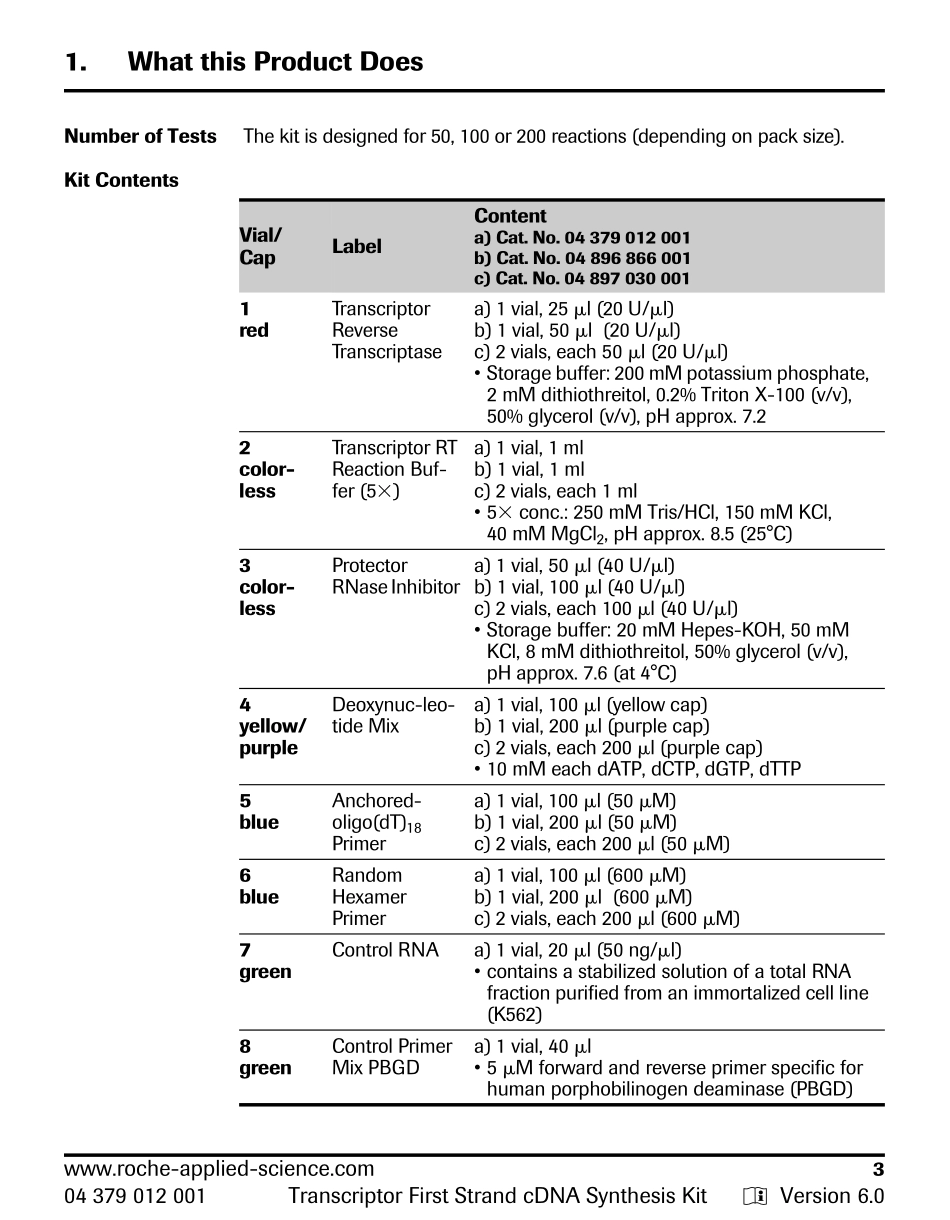
www.roche-applied-science.comForlifescienceresearchonly.Notforuseindiagnosticprocedures.TranscriptorFirstStrandcDNASynthesisKityVersion6.0Contentversion:September2010Cat.No.04379012001Kitfor50reactionsincluding10controlreactionsCat.No.04896866001Kitfor100reactionsCat.No.04897030001Kitfor200reactionsStorethekitat�15to�25°CNStoreControlRNA(vial7inCat.No04379012001)at—70°Corbelow.2www.roche-applied-science.com04379012001TranscriptorFirstStrandcDNASynthesisKityVersion6.0TableofContents1.WhatthisProductDoes........................................................................................................3NumberofTests3KitContents3StorageandStability4AdditionalEquipmentandReagentsRequired4Application42.HowtoUsethisProduct.......................................................................................................52.1BeforeYouBegin5Precautions5SampleMaterial5Primer62.2Procedure8StandardRT-PCRProcedure8ProcedureA:cDNASynthesiswithanchored-oligo(dT)18ORrandomORsequence-specificprimer8ProcedureB:cDNASynthesiswithanchored-oligo(dT)18primerANDrandomhexamerprimer112.3ControlReaction14cDNASynthesis14PCRforPBGD15PCRinaconventionalthermalblockcycler15PCRontheLightCycler®Carousel-BasedSystem16PCRontheLightCycler®480Instrument183.Results....................................................................................................................................204.Troubleshooting....................................................................................................................235.AdditionalInformationonthisProduct...........................................................................26HowthisProductWorks26References27QualityControl286.SupplementaryInformation...............................................................................................296.1Conventions29TextConventions29Symbols296.2ChangestoPreviousVersion296.3OrderingInformation296.4Trademarks31PROTOCOLPROTOCOLwww.roche-applied-science.com304379012001Transcr...