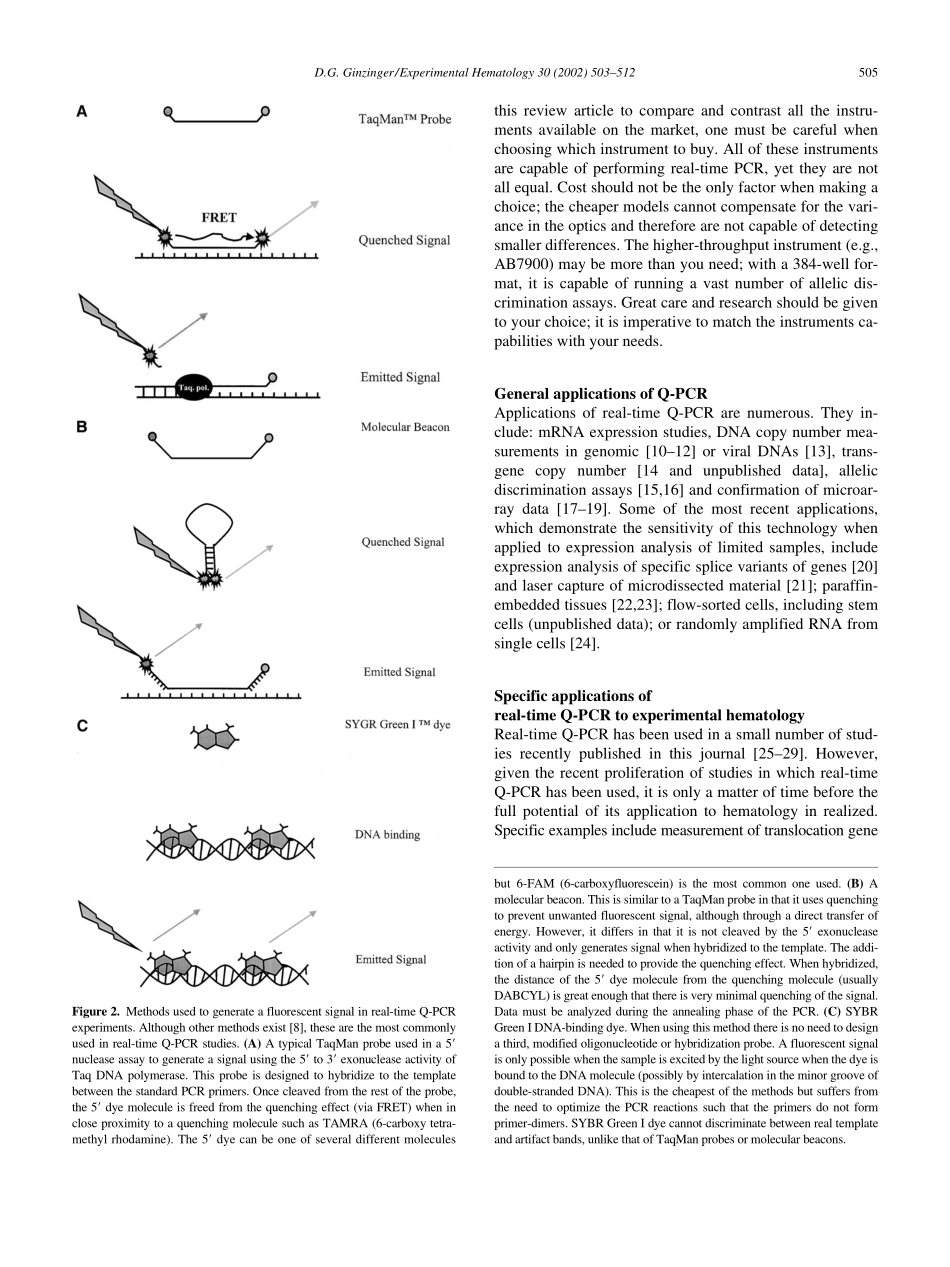
ExperimentalHematology30(2002)503–5120301-472X/02$–seefrontmatter.Copyright©2002InternationalSocietyforExperimentalHematology.PublishedbyElsevierScienceInc.PIIS0301-472X(02)00806-8Genequantificationusingreal-timequantitativePCR:AnemergingtechnologyhitsthemainstreamDavidG.GinzingerGenomeAnalysisCoreFacility,ComprehensiveCancerCenter,SchoolofMedicine,UniversityofCaliforniaatSanFrancisco,SanFrancisco,Calif.,USA(Received10December2001;revised20February2002;accepted5March2002)Therecentfloodofreportsusingreal-timeQ-PCRtestifiestothetransformationofthistech-nologyfromanexperimentaltoolintothescientificmainstream.Manyoftheapplicationsofreal-timeQ-PCRincludemeasuringmRNAexpressionlevels,DNAcopynumber,transgenecopynumberandexpressionanalysis,allelicdiscrimination,andmeasuringviraltiters.Therangeofapplicationsofreal-timeQ-PCRisimmenseandhasbeenfueledinpartbytheprolif-erationoflower-costinstrumentationandreagents.Successfulapplicationofreal-timeQ-PCRisnottrivial.However,thisreviewwillhelpguidethereaderthroughthevariablesthatcanlimittheusefulnessofthistechnology.Carefulconsiderationoftheassaydesign,templatepreparation,andanalyticalmethodsareessentialforaccurategenequantification.©2002In-ternationalSocietyforExperimentalHematology.PublishedbyElsevierScienceInc.Aliteraturesearchwiththekeyword“quantitativePCR(Q-PCR)”or“real-timePCR”willgenerateliterallythousandsofhits,testifyingtotheemergenceofthistechnologyintothemainstreamofmanyscientificdisciplines.Ifthesamesearchweretohavebeenperformedafewyearsago,onlyacoupleofhundredhitswouldhavebeenreturned.Whathasaccountedforthissuddenincreaseofpapersutilizingthistechnology?Thefirstdocumentationofreal-timePCRwasin1993[1],andyetthistechnologyhasonlyrecentlyhitthemainstream.Perhapstheprimaryreasonforthiswasthegreatexpenseoftheinstruments,butithascertainlybeenexacerbatedbythecomplexitiesofperformingreproduciblereal-timeQ-PCRstudies,andreal-timePCRhasonlyre-centlybeenwidelyacceptedasavaluabletechnique.Withtheincreasingnumberofreal-tim...