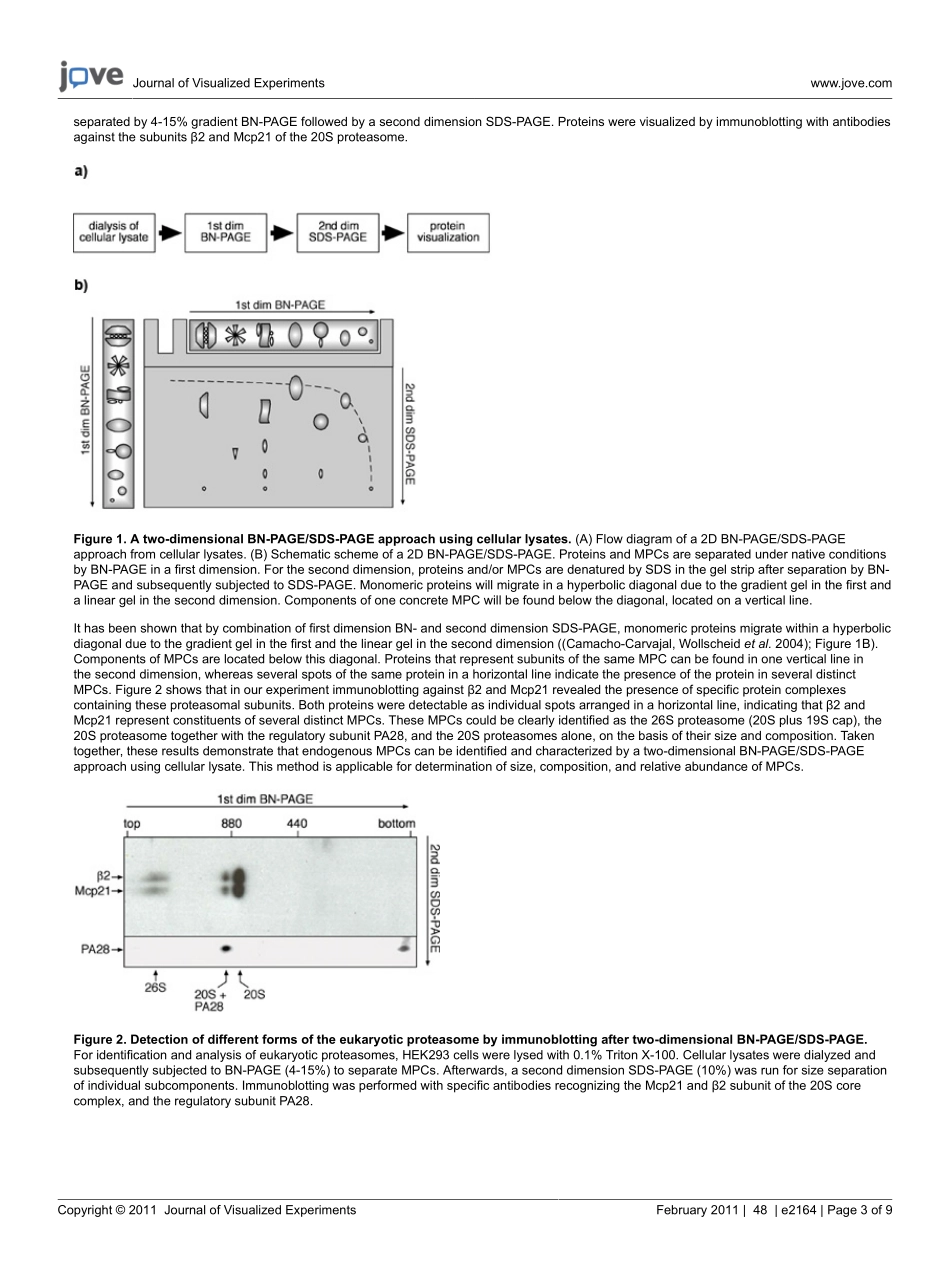
JournalofVisualizedExperimentswww.jove.comCopyright©2011JournalofVisualizedExperimentsFebruary2011|48|e2164|Page1of9VideoArticleBlueNativePolyacrylamideGelElectrophoresis(BN-PAGE)forAnalysisofMultiproteinComplexesfromCellularLysatesGinaJ.Fiala1,2,3,WolfgangW.A.Schamel*2,3,BrittaBlumenthal*31SpemannGraduateSchoolofBiologyandMedicine(SGBM),UniversityofFreiburg2CentreforBiologicalSignallingStudies(bioss)andBiologyIII,FacultyofBiology,UniversityofFreiburg3DepartmentofMolecularImmunology,Max-Planck-InstituteofImmunologyandEpigenetics*TheseauthorscontributedequallyCorrespondenceto:WolfgangW.A.Schamelatschamel@immunbio.mpg.de,BrittaBlumenthalatblumenthal@immunbio.mpg.deURL:http://www.jove.com/video/2164DOI:doi:10.3791/2164Keywords:Biochemistry,Issue48,BN-PAGE,2Dgelelectrophoresis,cellularlysate,dialysis,proteincomplex,multiproteincomplex,proteininteractionDatePublished:2/24/2011Citation:Fiala,G.J.,Schamel,W.W.A.,Blumenthal,B.BlueNativePolyacrylamideGelElectrophoresis(BN-PAGE)forAnalysisofMultiproteinComplexesfromCellularLysates.J.Vis.Exp.(48),e2164,doi:10.3791/2164(2011).AbstractMultiproteincomplexes(MPCs)playacrucialroleincellsignalling,sincemostproteinscanbefoundinfunctionalorregulatorycomplexeswithotherproteins(Sali,Glaeseretal.2003).Thus,thestudyofprotein-proteininteractionnetworksrequiresthedetailedcharacterizationofMPCstogainanintegrativeunderstandingofproteinfunctionandregulation.Foridentificationandanalysis,MPCsmustbeseparatedundernativeconditions.Inthisvideo,wedescribetheanalysisofMPCsbybluenativepolyacrylamidegelelectrophoresis(BN-PAGE).BN-PAGEisatechniquethatallowsseparationofMPCsinanativeconformationwithahigherresolutionthanofferedbygelfiltrationorsucrosedensityultracentrifugation,andisthereforeusefultodetermineMPCsize,composition,andrelativeabundance(SchäggerandvonJagow1991);(Schägger,Crameretal.1994).Bythismethod,proteinsareseparatedaccordingtotheirhydrodynamicsizeandshapeinapolyacrylamidematrix.Here,wedemonstratetheanalysisofMPCsoftotalcellularlysates,...