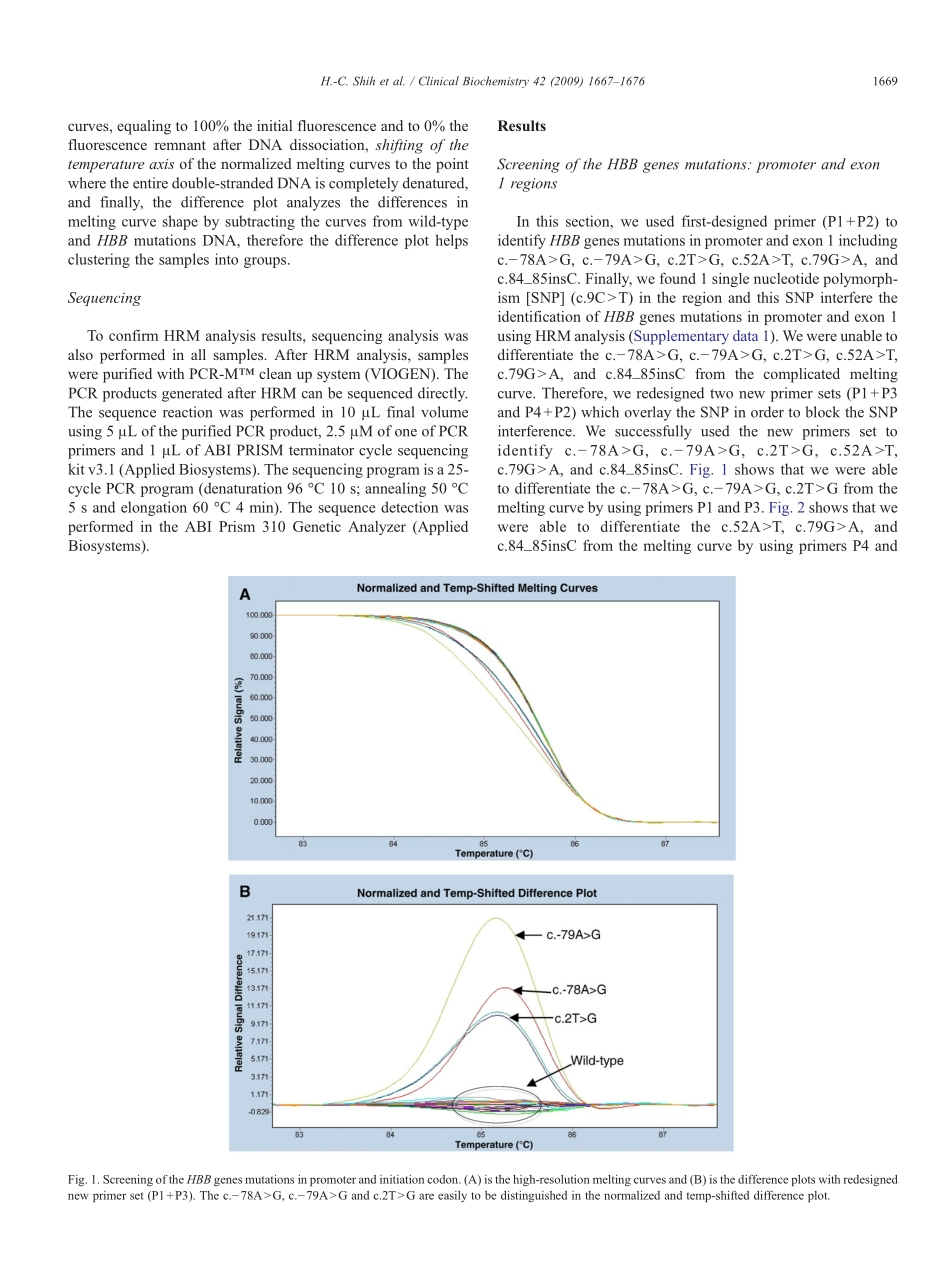
RapididentificationofHBBgenemutationsbyhigh-resolutionmeltinganalysisHung-ChangShiha,b,c,d,Tze-KiongErb,e,Tien-JyeChangc,Ya-SianChangb,Ta-ChihLiua,b,e,Jan-GowthChanga,b,e,f,⁎aInstituteofClinicalMedicine,CollegeofMedicine,KaohsiungMedicalUniversity,Kaohsiung,TaiwanbGraduateInstituteofMedicine,CollegeofMedicine,KaohsiungMedicalUniversity,Kaohsiung,TaiwancDepartmentofVeterinaryMedicine,NationalChungHsiungUniversity,Taichung,TaiwandDepartmentofLaboratoryMedicine,ChinaMedicalUniversityHospital,Taichung,TaiwaneDepartmentofLaboratoryMedicine,KaohsiungMedicalUniversityHospital,Kaohsiung,TaiwanfCenterforExcellenceinEnvironmentalMedicine,KaohsiungMedicalUniversity,Kaohsiung,TaiwanReceived24April2009;receivedinrevisedform3July2009;accepted14July2009Availableonline23July2009AbstractObjective:ThisstudywasundertakentoidentifyHBBgenemutation.Designandmethods:Hereinweevaluatedhigh-resolutionmeltinganalysisintheidentificationofHBBmutations.Results:WehavesuccessfullyestablishedadiagnosticstrategyforidentifyingHBBgenemutationsincludingc.−78ANG,c.−79ANG,c.2TNG,c.79_80insT,c.84_85insC,c.123_124insT,c.125_128delTCTT,c.130GNT,c.170GNA,c.216_217insAandc.316–197CNTfromwild-typeDNAusingHRManalysis.TheresultsofHRManalysiswereconfirmedbydirectDNAsequencing.Conclusions:Insummary,wereportthatHRManalysisisanappealingtechniquefortheidentificationofHBBmutations.WealsobelievethatHRMcanbeusedasamethodforprenataldiagnosisofβ-thalassemia.©2009TheCanadianSocietyofClinicalChemists.PublishedbyElsevierInc.Allrightsreserved.Keywords:HBBgene;Mutation;β-thalassemia;High-resolutionmeltinganalysis;Singlenucleotidepolymorphism;MeltingcurveIntroductionHemoglobinopathiesresultingfrommutationsintheα-orβ-likeglobingeneclustersarethemostcommoninheriteddisordersinhumans,witharound7%oftheworldpopulationbeingcarriersofaglobingenemutation[1].Moleculardefectsineitherregulatoryorcodingregionsofthehumanα-,β-orδ-globingenescanminimallyordrasticallyreducetheirexpression,leadingtoα-,β-orδ-thalassemia,respecti...