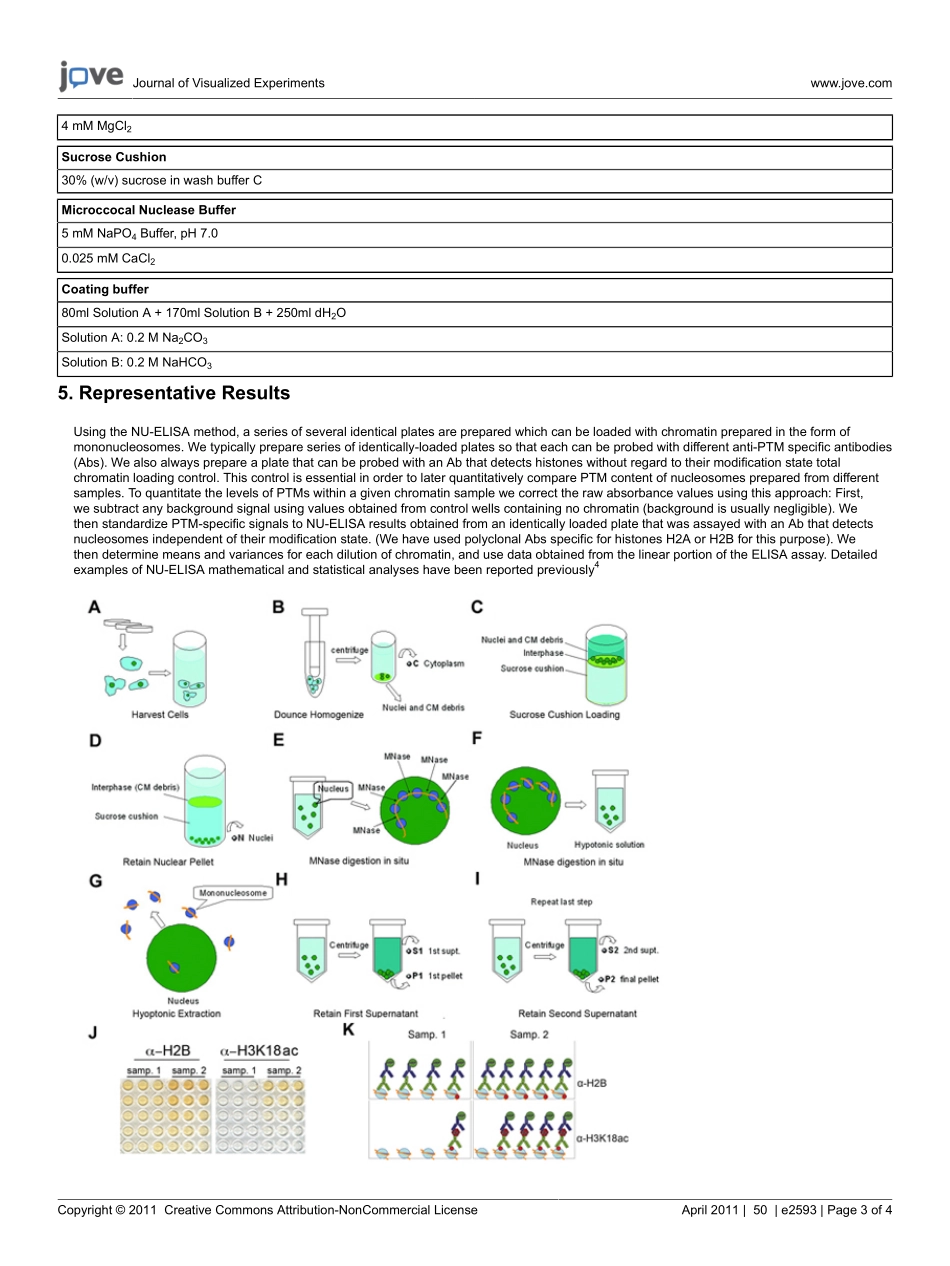
JournalofVisualizedExperimentswww.jove.comCopyright©2011CreativeCommonsAttribution-NonCommercialLicenseApril2011|50|e2593|Page1of4VideoArticleDetectionofPost-translationalModificationsonNativeIntactNucleosomesbyELISABoDai1,FaridaDahmani2,JosephA.Cichocki3,LindseyC.Swanson2,TheodoreP.Rasmussen31InstituteforStemCellBiologyandRegenerativeMedicine,StanfordUniversity2DepartmentofMolecularandCellBiology,UniversityofConnecticut3DepartmentofPharmaceuticalSciences,UniversityofConnecticutCorrespondenceto:TheodoreP.Rasmussenattheodore.rasmussen@uconn.eduURL:http://www.jove.com/video/2593DOI:doi:10.3791/2593Keywords:CellularBiology,Issue50,Chromatin,Nucleosome,Epigenetics,ELISA,Histone,Modification,Methylation,AcetylationDatePublished:4/26/2011Citation:Dai,B.,Dahmani,F.,Cichocki,J.A.,Swanson,L.C.,Rasmussen,T.P.DetectionofPost-translationalModificationsonNativeIntactNucleosomesbyELISA.J.Vis.Exp.(50),e2593,doi:10.3791/2593(2011).AbstractThegenomeofeukaryotesexistsaschromatinwhichcontainsbothDNAandproteins.Thefundamentalunitofchromatinisthenucleosome,whichcontains146basepairsofDNAassociatedwithtwoeachofhistonesH2A,H2B,H3,andH41.TheN-terminaltailsofhistonesarerichinlysineandarginineandaremodifiedpost-transcriptionallybyacetylation,methylation,andotherpost-translationalmodifications(PTMs).ThePTMconfigurationofnucleosomescanaffectthetranscriptionalactivityofassociatedDNA,thusprovidingamodeofgeneregulationthatisepigeneticinnature2,3.WedevelopedamethodcallednucleosomeELISA(NU-ELISA)toquantitativelydetermineglobalPTMsignaturesofnucleosomesextractedfromcells.NU-ELISAismoresensitiveandquantitativethanwesternblotting,andisusefultointerrogatetheepiproteomicstateofspecificcelltypes.ThisvideojournalarticleshowsdetailedprocedurestoperformNU-ELISAanalysis.VideoLinkThevideocomponentofthisarticlecanbefoundathttp://www.jove.com/video/2593/Protocol1.MammalianCellCultureNU-ELISAcanbeperformedonanymammaliancelltypethatcanbegrowninculture.Weprefertopreparemoderatetolargebatchesofcellssothatnucleos...