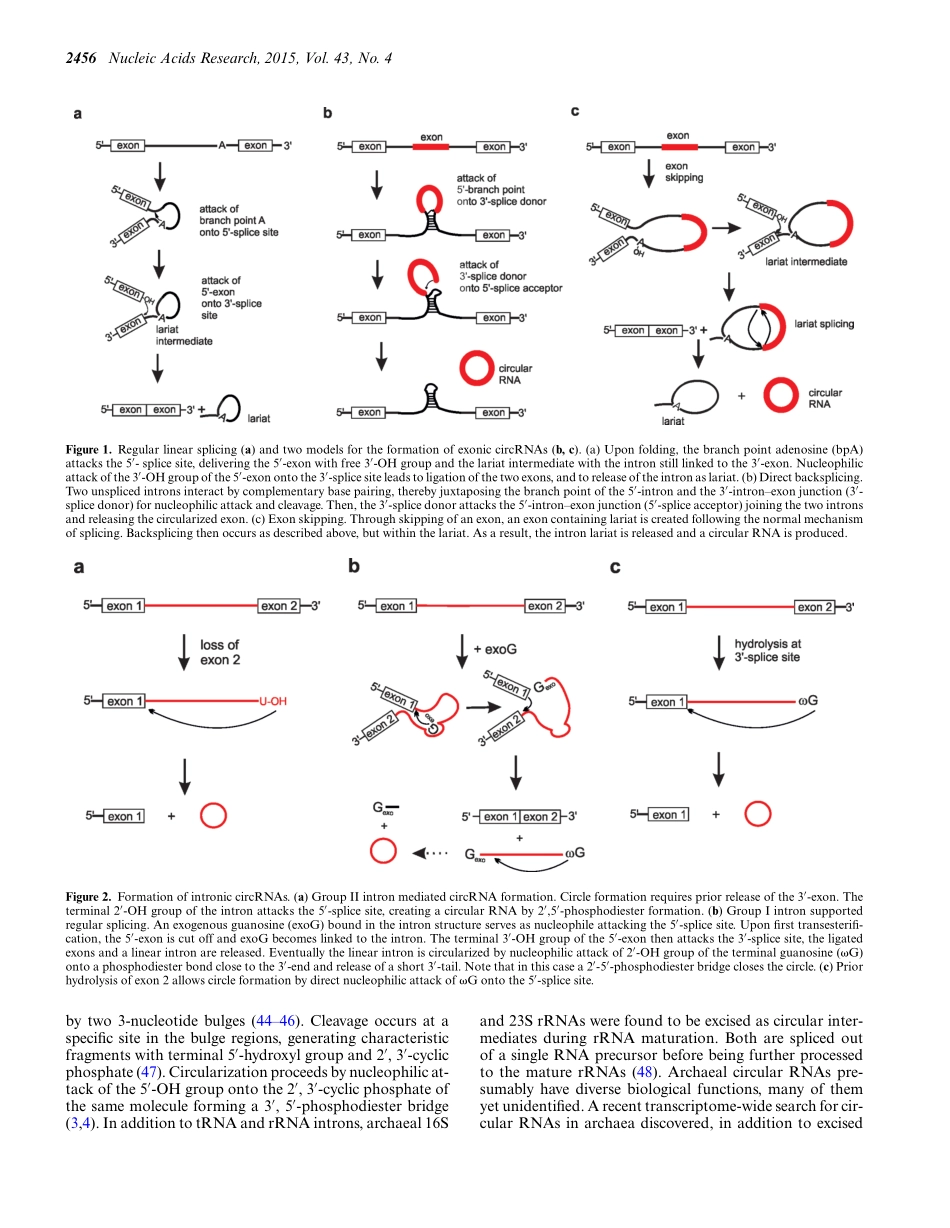
2454–2465NucleicAcidsResearch,2015,Vol.43,No.4Publishedonline6February2015doi:10.1093/nar/gkv045SURVEYANDSUMMARYRNAcircularizationstrategiesinvivoandinvitroSonjaPetkovicandSabineM¨uller*Institutf¨urBiochemie,ErnstMoritzArndtUniversit¨atGreifswald,Felix-Hausdorff-Str.4,17487Greifswald,GermanyReceivedMarch26,2014;RevisedJanuary07,2015;AcceptedJanuary12,2015ABSTRACTIntheplenitudeofnaturallyoccurringRNAs,circularRNAs(circRNAs)andtheirbiologicalrolewereun-derestimatedforyears.However,circRNAsareubiq-uitousinalldomainsoflife,includingeukaryotes,archaea,bacteriaandviruses,wheretheycanful-filldiversebiologicalfunctions.Someofthosefunc-tions,asforexampleplayingaroleinthelifecy-cleofviralandviroidgenomesorinthematurationoftRNAgenes,havebeenelucidated;otherputativefunctionsstillremainelusive.Duetotheresistancetoexonucleases,circRNAsarepromisingtoolsforinvivoapplicationasaptamers,trans-cleavingri-bozymesorsiRNAs.HowarecircRNAsgeneratedinvivoandwhatapproachesdoexisttoproducering-shapedRNAsinvitro?InthisreviewweillustratetheoccurrenceandmechanismsofRNAcircularizationinvivo,surveymethodsforthegenerationofcircRNAinvitroandprovideappropriateprotocols.INTRODUCTIONCircularRNAsarefoundinallkingdomsoflife,appearingforexampleasgenomesofviroidalplantpathogens(1)andofthehepatitisdeltavirus(HDV)(2),orassplicedtRNAandrRNAintronsandasrRNAprocessingintermediatesinarchaea(3,4).Furthermore,circRNAsareformedinthelifecycleofbacterialandeukaryalgroupIintrons,wheretheyweresuggestedtoplayaroleinintronmobilitybyre-versesplicing(5–7).However,circRNAswereconsideredextremelyrareinnaturefordecades,andinparticularineukaryotes,circRNAswereseenasminorRNAstructuralvariantsattributedtotranscriptionalnoise(8).Thisviewhasdramaticallychanged,asanumberofrecentreportshaveconvincinglydemonstratedthatcircRNAsineukary-otesarehighlyabundantandevolutionaryconserved.Ap-parently,thousandsofhumantranscriptsareexpressedascircularisoformsoftheirlinearcounterparts(9–13).ThefunctionsofcircRNAsineukaryotesstillremainmo...