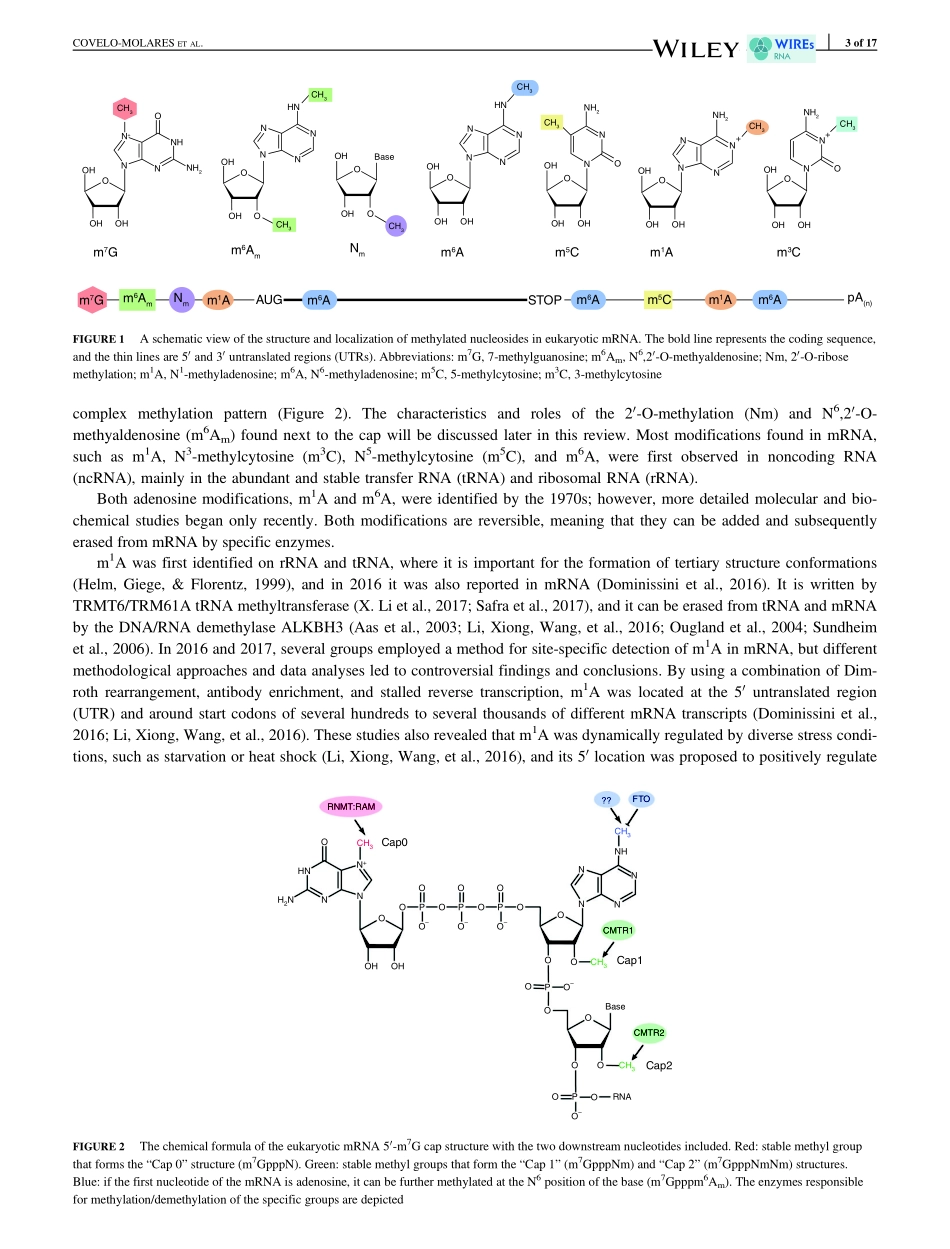
ADVANCEDREVIEWRNAmethylationinnuclearpre-mRNAprocessingHelenaCovelo-Molares|MarekBartosovic†|StepankaVanacovaCEITEC,MasarykUniversity,Brno,CzechRepublicCorrespondenceStepankaVanacova,CEITEC,MasarykUniversity,Brno,CzechRepublic.Email:stepanka.vanacova@ceitec.muni.cz†PresentaddressDepartmentofMedicalBiochemistryandBiophysics,KarolinskaInstitutet,Solna,Sweden.FundinginformationBrnoCityMunicipalityScholarship;MinistryofEducation,YouthandSports,Grant/AwardNumber:LQ1601;CzechScienceFoundation,Grant/AwardNumbers:305/11/1095,14-25884S;WellcomeTrust,Grant/AwardNumber:084316/B/07/ZEukaryoticRNAcancarrymorethan100differenttypesofchemicalmodifica-tions.EarlystudieshavebeenfocusedonmodificationsofhighlyabundantRNA,suchasribosomalRNAandtransferRNA,butrecenttechnicaladvanceshavemadeitpossibletoalsostudymessengerRNA(mRNA).Subsequently,mRNAmodifications,namelymethylation,haveemergedaskeyplayersineukaryoticgeneexpressionregulation.ThemostabundantandwidelystudiedinternalmRNAmodificationisN6-methyladenosine(m6A),butthelistofmRNAchemicalmodifi-cationscontinuestogrowasfastasinterestinthisfield.Overthepastdecade,transcriptome-widestudiescombinedwithadvancedbiochemistryandthediscov-eryofmethylationwriters,readers,anderasersrevealedrolesformRNAmethyla-tionintheregulationofnearlyeveryaspectofthemRNAlifecycleandindiversecellular,developmental,anddiseaseprocesses.AlthoughlargepartsofmRNAfunctionarelinkedtoitscytoplasmicstabilityandregulationofitstranslation,anumberofstudieshavebeguntoprovideevidenceformethylation-regulatednuclearprocesses.Inthisreview,wesummarizetherecentadvancesinRNAmeth-ylationresearchandhighlighthowthesenewfindingshavecontributedtoourunderstandingofmethylation-dependentRNAprocessinginthenucleus.Thisarticleiscategorizedunder:RNAProcessing>RNAEditingandModificationRNAProcessing>SplicingRegulation/AlternativeSplicingRNAInteractionswithProteinsandOtherMolecules>Protein–RNAInterac-tions:FunctionalImplicationsKEYWORDSRNAdemethylase,RNAmethylase,RNAprocessing1|INTRODUCTIONPo...