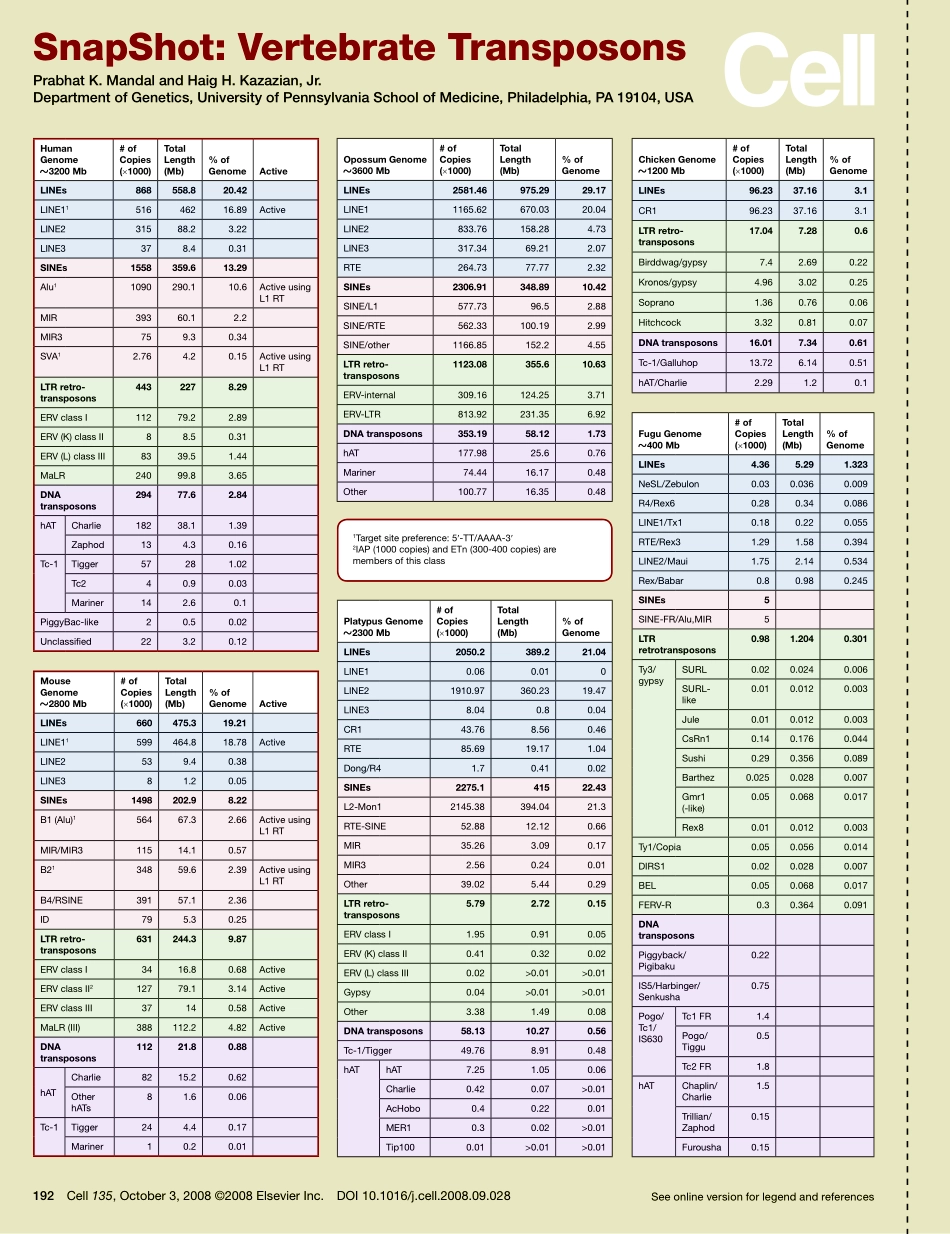
Seeonlineversionforlegendandreferences192Cell135,October3,2008©2008ElsevierInc.DOI10.1016/j.cell.2008.09.028SnapShot:VertebrateTransposonsPrabhatK.MandalandHaigH.Kazazian,Jr.DepartmentofGenetics,UniversityofPennsylvaniaSchoolofMedicine,Philadelphia,PA19104,USAHumanGenome?3200Mb#ofCopies(×1000)TotalLength(Mb)%ofGenomeActiveLINEs868558.820.42LINE1151646216.89ActiveLINE231588.23.22LINE3378.40.31SINEs1558359.613.29Alu11090290.110.6ActiveusingL1RTMIR39360.12.2MIR3759.30.34SVA12.764.20.15ActiveusingL1RTLTRretrotransposons4432278.29ERVclassI11279.22.89ERV(K)classII88.50.31ERV(L)classIII8339.51.44MaLR24099.83.65DNAtransposons29477.62.84hATCharlie18238.11.39Zaphod134.30.16Tc-1Tigger57281.02Tc240.90.03Mariner142.60.1PiggyBac-like20.50.02Unclassified223.20.12OpossumGenome?3600Mb#ofCopies(×1000)TotalLength(Mb)%ofGenomeLINEs2581.46975.2929.17LINE11165.62670.0320.04LINE2833.76158.284.73LINE3317.3469.212.07RTE264.7377.772.32SINEs2306.91348.8910.42SINE/L1577.7396.52.88SINE/RTE562.33100.192.99SINE/other1166.85152.24.55LTRretrotransposons1123.08355.610.63ERV-internal309.16124.253.71ERV-LTR813.92231.356.92DNAtransposons353.1958.121.73hAT177.9825.60.76Mariner74.4416.170.48Other100.7716.350.48PlatypusGenome?2300Mb#ofCopies(×1000)TotalLength(Mb)%ofGenomeLINEs2050.2389.221.04LINE10.060.010LINE21910.97360.2319.47LINE38.040.80.04CR143.768.560.46RTE85.6919.171.04Dong/R41.70.410.02SINEs2275.141522.43L2-Mon12145.38394.0421.3RTE-SINE52.8812.120.66MIR35.263.090.17MIR32.560.240.01Other39.025.440.29LTRretrotransposons5.792.720.15ERVclassI1.950.910.05ERV(K)classII0.410.320.02ERV(L)classIII0.02>0.01>0.01Gypsy0.04>0.01>0.01Other3.381.490.08DNAtransposons58.1310.270.56Tc-1/Tigger49.768.910.48hAThAT7.251.050.06Charlie0.420.07>0.01AcHobo0.40.220.01MER10.30.02>0.01Tip1000.01>0.01>0.01ChickenGenome?1200Mb#ofCopies(×1000)TotalLength(Mb)%ofGenomeLINEs96.2337.163.1CR196.2337.163.1LTRretrotransposons17.047.280.6Birddwag/gypsy7.42.690.22Kronos/gypsy4.963.020.25Soprano1.360.760.06Hitchcock3.320....