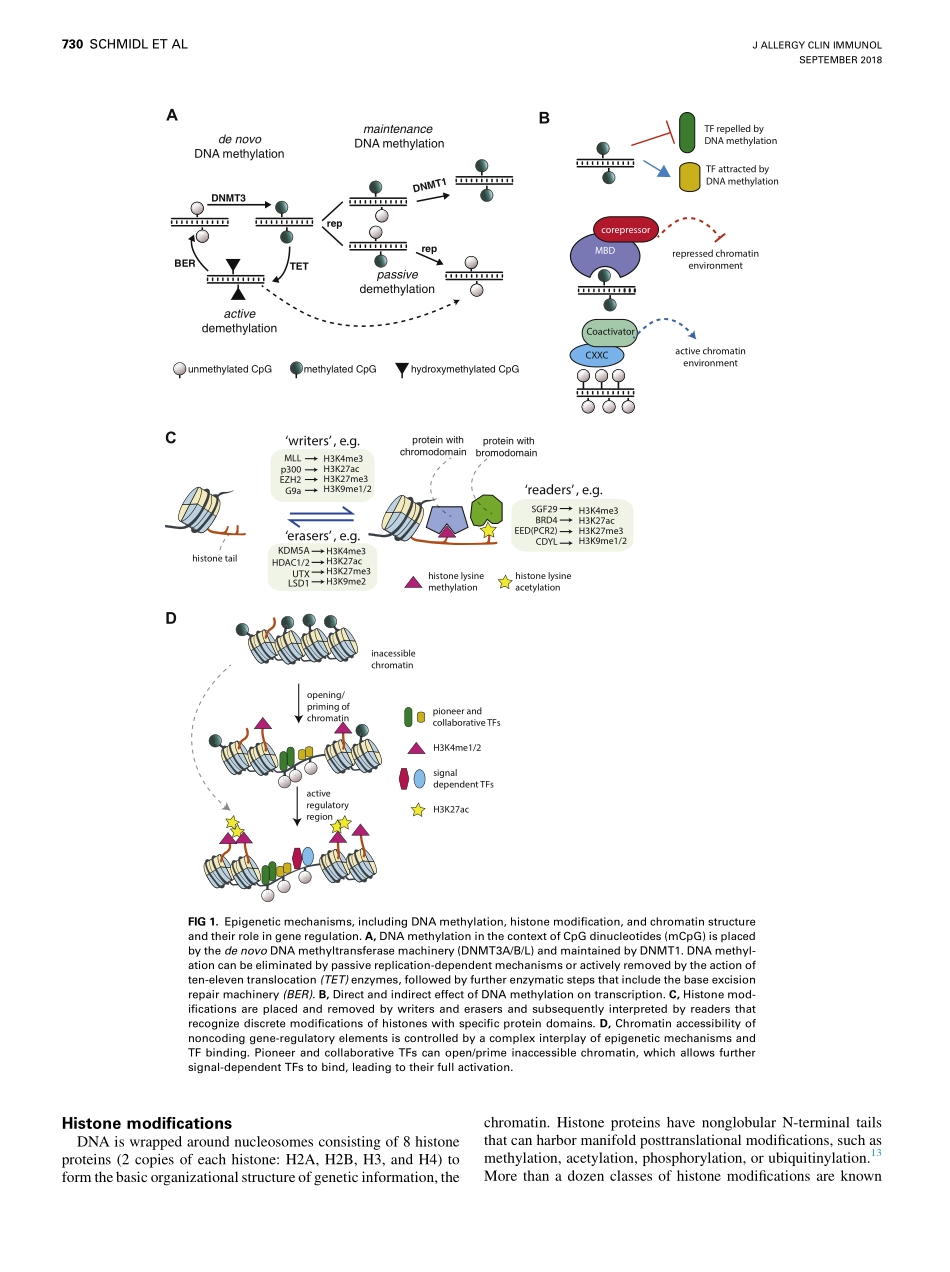
MechanismsofallergicdiseasesEpigeneticmechanismsregulatingT-cellresponsesChristianSchmidl,PhD,aMichaelDelacher,PhD,a,bJochenHuehn,PhD,candMarkusFeuerer,MDa,bRegensburgandBraunschweig,GermanyDuringthelastdecade,advancesinsequencingtechnologiesallowedproductionofawealthofinformationonepigeneticmodificationsinTcells.Epigenomemaps,incombinationwithmechanisticstudies,havedemonstratedthatTcellsundergoextensiveepigenomeremodelinginresponsetosignals,whichhasastrongeffectonphenotypicstabilityandfunctionoflymphocytes.InthisreviewwefocusonDNAmethylation,histonemodifications,andchromatinstructureasimportantepigeneticmechanismsinvolvedincontrollingT-cellresponses.Inparticular,wediscussepigeneticprocessesinlightofthedevelopment,activation,anddifferentiationofCD41Thelper(TH),regulatoryT,andCD81Tcells.Ascentralaspectsoftheadaptiveimmunesystem,wereviewmechanismsthatensuremolecularmemory,stability,plasticity,andexhaustionofTcells.WefurtherdiscusstheeffectofthetissueenvironmentonimprintingT-cellepigenomeswithpotentialimplicationsforimmunotherapy.(JAllergyClinImmunol2018;142:728-43.)Keywords:Epigenetics,generegulation,enhancer,promoter,chro-matinaccessibility,histonemodifications,DNAmethylation,tran-scriptionfactorbinding,T-celldevelopment,T-cellfunction,T-cellexhaustion,regulatoryTcells,tissuespecificityTcellsareasubtypeofwhitebloodcells,whichplayamajorroleintheadaptiveimmunesystem.Theycanbeclassifiedaccordingtotheirspecificfunction:MHCclassII–restrictedCD4-expressingTcells(CD4)candifferentiateintospecificTHsubtypes.Incontrasttothis,MHCclassI–restrictedCD8-expressingTcells(CD8)areclassicallyknownfortheirpotentialtodestroyvirus-infectedortumorcells,hencetheirdenominationascytotoxicTlymphocytes(CTLs).FollowingdevelopmentinthethymusfromearlyT-cellprecursorcellsoverCD4andCD8double-negative(DN)anddouble-positive(DP)stages,TcellsfinallydifferentiateintoCD4andCD8single-positive(SP)thy-mocytes.OnrecognizingMHC-peptidecomplexesthroughtheirT-cellreceptor(TCR),naiveCD41orCD81Tcellscandiffer...