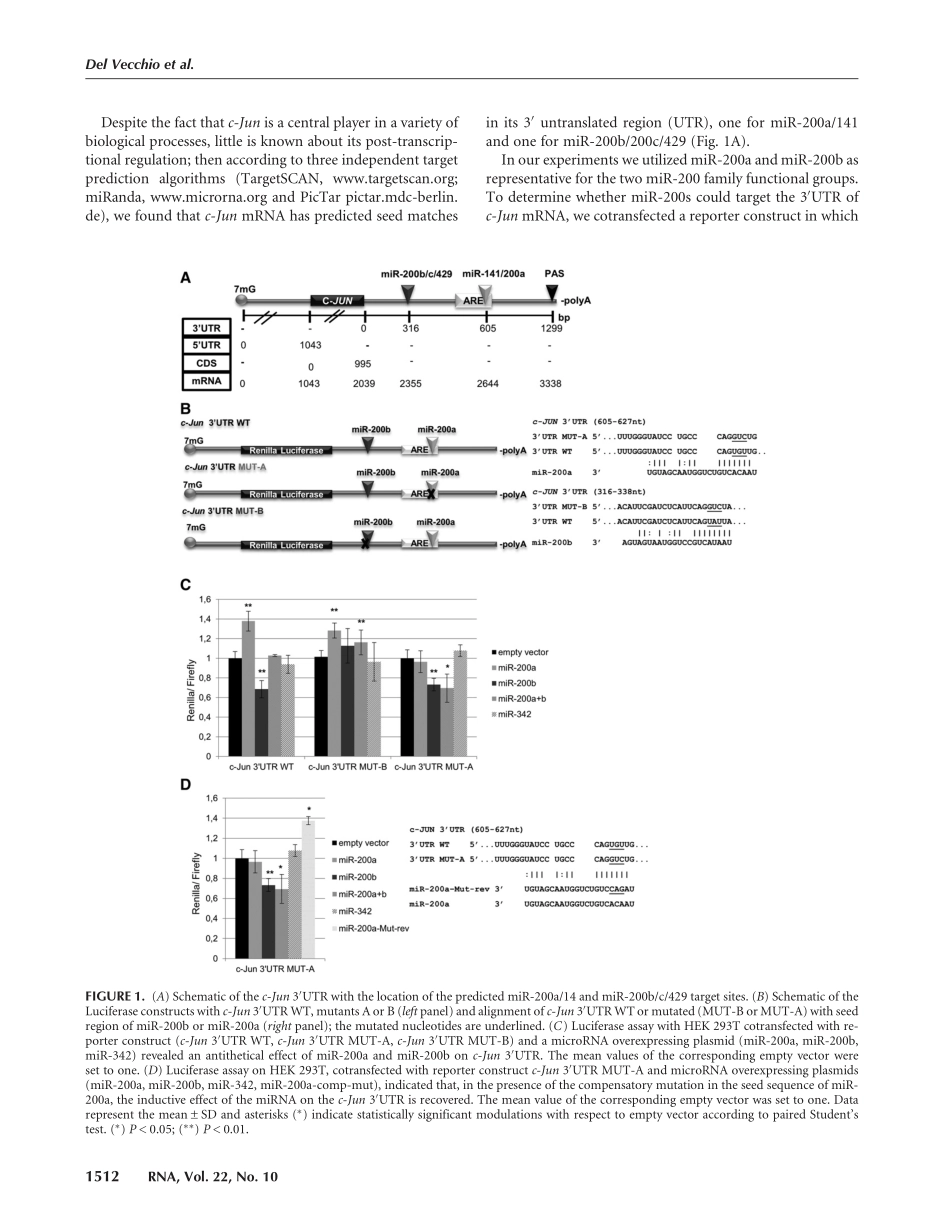
RNA-bindingproteinHuRandthemembersofthemiR-200familyplayanunconventionalroleintheregulationofc-JunmRNAGIORGIADELVECCHIO,1,4FRANCESCADEVITO,1,4SITAJ.SAUNDERS,2ADELERISI,1CECILIAMANNIRONI,3IRENEBOZZONI,1andCARLOPRESUTTI11DipartimentodiBiologiaeBiotecnologie,Università“Sapienza,”00185Rome,Italy2BioinformaticsGroup,DepartmentofComputerScience,Albert-Ludwigs-UniversityFreiburg,79110Freiburg,Germany3IBPM–CNR,00185Rome,ItalyABSTRACTPost-transcriptionalgeneregulationisafundamentalstepforcoordinatingcellularresponseinavarietyofprocesses.RNA-bindingproteins(RBPs)andmicroRNAs(miRNAs)arethemostimportantfactorsresponsibleforthisregulation.HerewereportthatdifferentcomponentsofthemiR-200familyareinvolvedinc-JunmRNAregulationwiththeoppositeeffect.WhilemiR-200binhibitsc-Junproteinproduction,miR-200atendstoincreasetheJUNamountthroughastabilizationofitsmRNA.ThisactionisdependentonthepresenceoftheRBPHuRthatbindsthe3′UTRofc-JunmRNAinaregionincludingthemir-200abindingsite.Thepositionofthebindingsiteisfundamental;bymutatingthissite,wedemonstratethattheeffectisnotmicro-RNAspecific.TheseresultsindicatethatmiR-200atriggersamicroRNA-mediatedstabilizationofc-JunmRNA,promotingthebindingofHuRwithc-JunmRNA.ThisisthefirstexampleofapositiveregulationexertedbyamicroRNAonanimportantoncogeneinproliferatingcells.Keywords:miRNA;HuR;mRNAstability;c-JunINTRODUCTIONPost-transcriptionalgeneregulationiscrucialtomaintainafineandcoordinatedproductionofproteinsnecessarytothecellsinavarietyofprocesseslikedevelopment,homeosta-sis,anddisease.Toachievethisresult,post-transcriptionalgeneexpressioniscontrolledatmultiplelevels:pre-mRNAsplicingandmat-uration,mRNAstabilityinthenucleus,mRNAtransport,editing,mRNAstabilityinthecytoplasm,storage,andtrans-lation(Bousquet-Antonellietal.2000;MitchellandToller-vey2000;OrphanidesandReinberg2000).mRNAturnoverandtranslationareverywell-suitedstepsofcontroltoaccom-plishquickchangesinthepatternofexpressedproteinsfol-lowingenvironmentalperturbations.Themostimportantcellularfac...