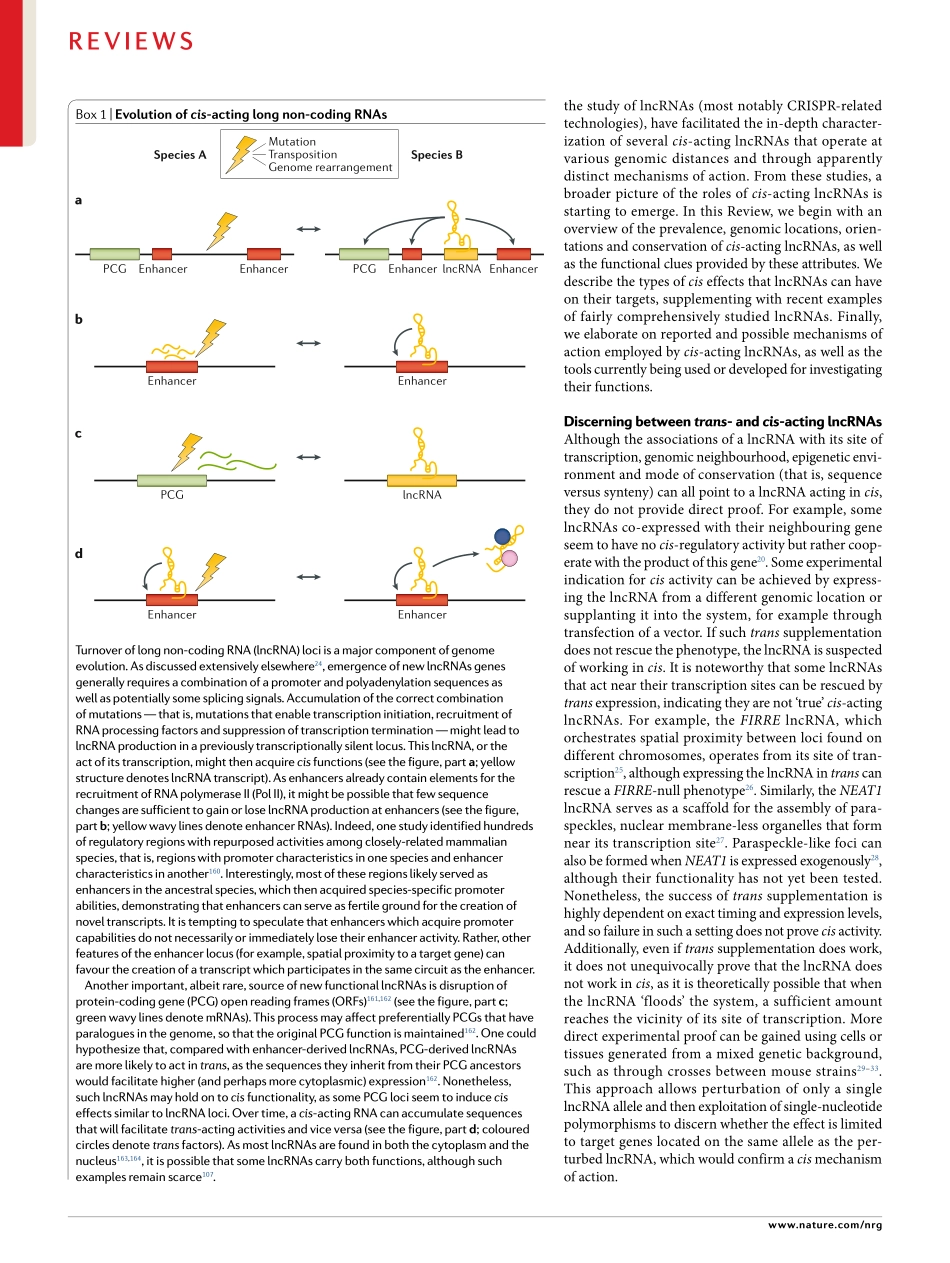
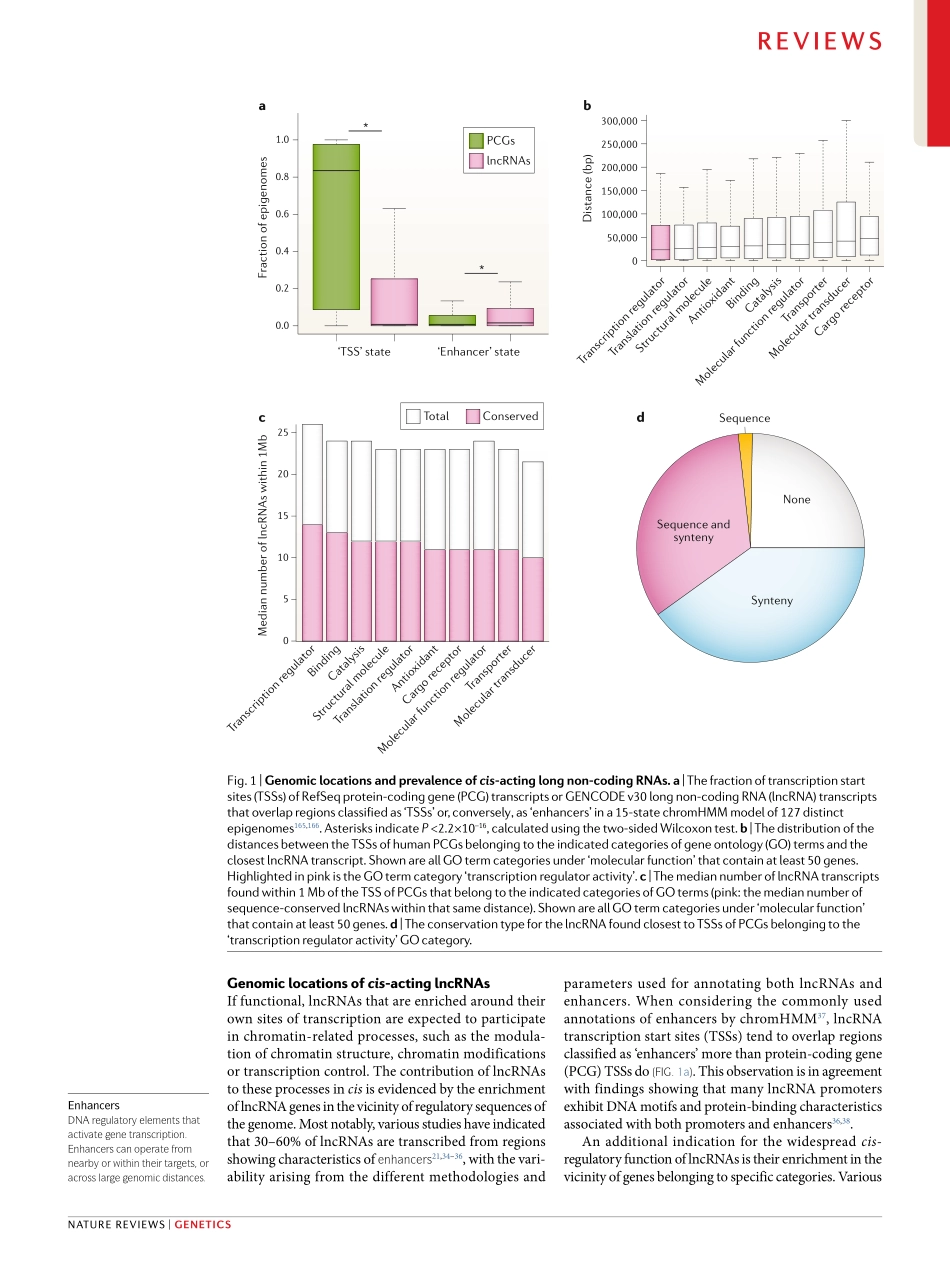
High-throughputsequencingtechnologiesandchroma-tinstatemapshaveshownthateukaryoticcellsproduceaplethoraofnon-codingtranscripts1–3.Ofthese,longnon-codingRNAs(lncRNAs)aredefinedarbitrarilyastranscriptsofmorethan200nucleotidesthatdonotbelongtoanyotherwell-definedgroupofnon-codingRNAs,suchasribosomalRNAs.Throughvariousmechanisms,lncRNAshavebeenimplicatedinawidearrayofcellularprocesses,includingtranscriptionalregulation,differentiation,cellularreprogrammingandmanyothers(reviewedelsewhere4–6).Withvaryinglevelsofevidence,lncRNAshavealsobeenimplicatedinvari-oushumandiseases7–9.lncRNAsaretranscribedbyRNApolymeraseII(PolII),andtheirbiogenesisissimilartomRNAsinthattheyarecappedandpolyadenylated.lncRNAsarealsousuallyspliced,althoughtheirexonnumberandsplicingefficiencyareonaveragelowerthanthoseofmRNAs10–13.However,aslncRNAsarepredominantlydefinedbyexclusioncriteria,thesetofgenesannotatedaslncRNAsincludesmanydistinctsub-groups,exemplifyingdiversestructuraland,presumably,functionalcharacteristics.AssigninglncRNAstodistinctfunctionalgroupsisessentialtoidentifycommonprin-ciples,andthuscomprisesapivotalstepwhenbeginningtoelucidatetheirroles.Thisstepremainsverychalleng-ing,withlimitedprogressbeingmadeinthepastdecadeoflncRNAresearch.OnetypeoflncRNAclassificationisbasedonthelocationatwhichthelncRNAfunctionsrelativetoitstranscriptionsite.Trans-actinglncRNAsaretranscribed,processedandthenvacatetheirsitesoftranscriptiontoexerttheirfunctionelsewhere,akintomRNAs.Theirfinaldestination,beitinthecytoplasmornucleus,doesnotdependontheirtranscriptionsite.Accordingly,aslongastheirlevelsareproperlymaintained,transcrib-ingtheselncRNAsfromadifferentgenomiclocationorsupplantingthemintothesystemshouldnotinterferewiththeirfunction(thatis,theirlossoffunctioncanberescuedbytheirexpressionfromexogenouslocations).AfewexamplesofsuchlncRNAshavenowbeenexten-sivelycharacterized14–16,andmanyadditionallncRNAshavebeenascribedtransactivities17–20.Bycontrast,cis-actinglncRNAsarethosewhoseactivityisbasedatandd...