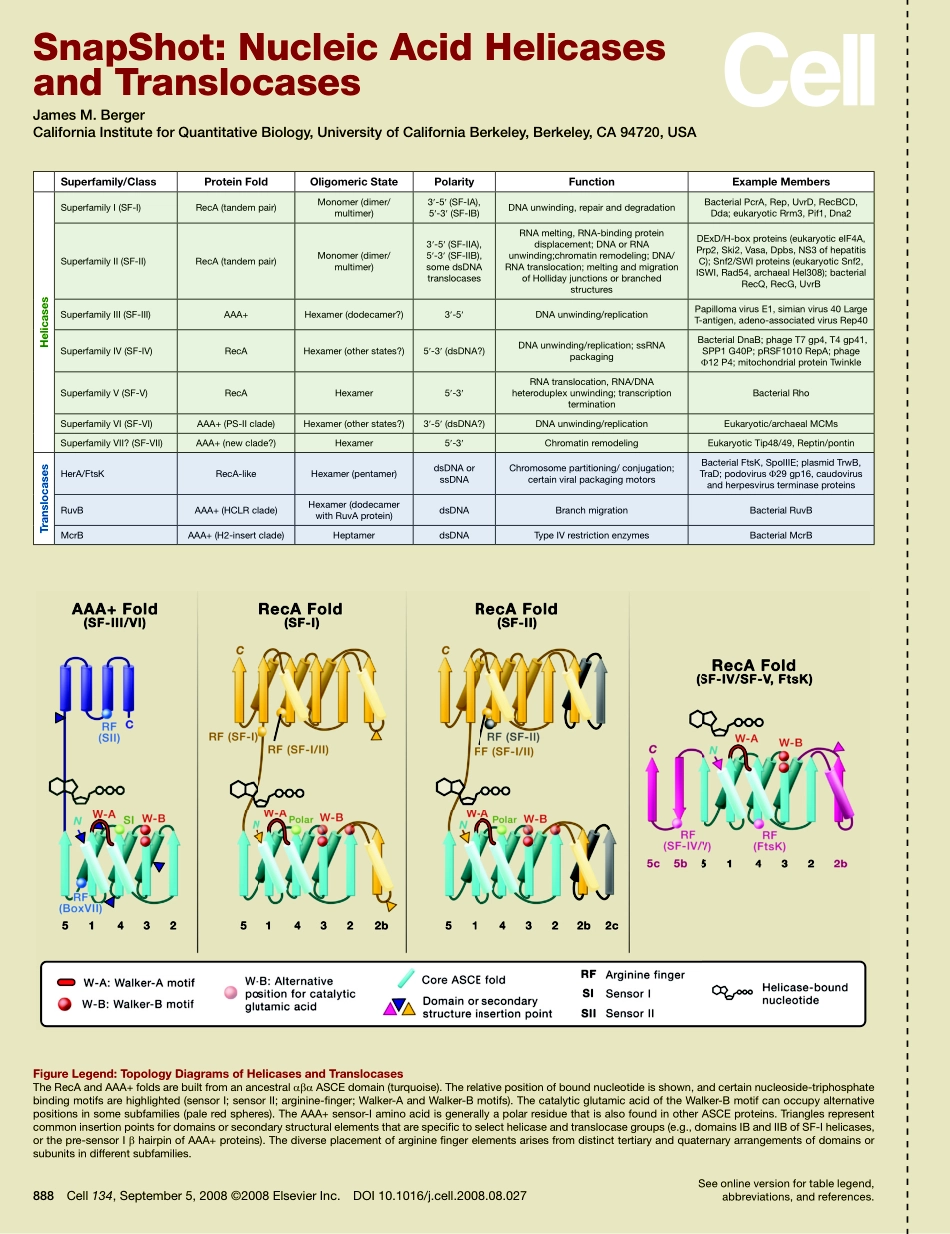
888Cell134,September5,2008©2008ElsevierInc.DOI10.1016/j.cell.2008.08.027Seeonlineversionfortablelegend,abbreviations,andreferences.SnapShot:NucleicAcidHelicasesandTranslocasesJamesM.BergerCaliforniaInstituteforQuantitativeBiology,UniversityofCaliforniaBerkeley,Berkeley,CA94720,USAsuperfamily/classProteinfoldoligomericstatePolarityfunctionexamplemembersHelicasesSuperfamilyI(SF-I)RecA(tandempair)Monomer(dimer/multimer)3′-5′(SF-IA),5′-3′(SF-IB)DNAunwinding,repairanddegradationBacterialPcrA,Rep,UvrD,RecBCD,Dda;eukaryoticRrm3,Pif1,Dna2SuperfamilyII(SF-II)RecA(tandempair)Monomer(dimer/multimer)3′-5′(SF-IIA),5′-3′(SF-IIB),somedsDNAtranslocasesRNAmelting,RNA-bindingproteindisplacement;DNAorRNAunwinding;chromatinremodeling;DNA/RNAtranslocation;meltingandmigrationofHollidayjunctionsorbranchedstructuresDExD/H-boxproteins(eukaryoticeIF4A,Prp2,Ski2,Vasa,Dpbs,NS3ofhepatitisC);Snf2/SWIproteins(eukaryoticSnf2,ISWI,Rad54,archaealHel308);bacterialRecQ,RecG,UvrBSuperfamilyIII(SF-III)AAA+Hexamer(dodecamer?)3′-5′DNAunwinding/replicationPapillomavirusE1,simianvirus40LargeT-antigen,adeno-associatedvirusRep40SuperfamilyIV(SF-IV)RecAHexamer(otherstates?)5′-3′(dsDNA?)DNAunwinding/replication;ssRNApackagingBacterialDnaB;phageT7gp4,T4gp41,SPP1G40P;pRSF1010RepA;phageΦ12P4;mitochondrialproteinTwinkleSuperfamilyV(SF-V)RecAHexamer5′-3′RNAtranslocation,RNA/DNAheteroduplexunwinding;transcriptionterminationBacterialRhoSuperfamilyVI(SF-VI)AAA+(PS-IIclade)Hexamer(otherstates?)3′-5′(dsDNA?)DNAunwinding/replicationEukaryotic/archaealMCMsSuperfamilyVII?(SF-VII)AAA+(newclade?)Hexamer5′-3′ChromatinremodelingEukaryoticTip48/49,Reptin/pontintranslocasesHerA/FtsKRecA-likeHexamer(pentamer)dsDNAorssDNAChromosomepartitioning/conjugation;certainviralpackagingmotorsBacterialFtsK,SpoIIIE;plasmidTrwB,TraD;podovirusΦ29gp16,caudovirusandherpesvirusterminaseproteinsRuvBAAA+(HCLRclade)Hexamer(dodecamerwithRuvAprotein)dsDNABranchmigrationBacterialRuvBMcrBAAA+(H2-insertclade)HeptamerdsDNATypeIVrestr...