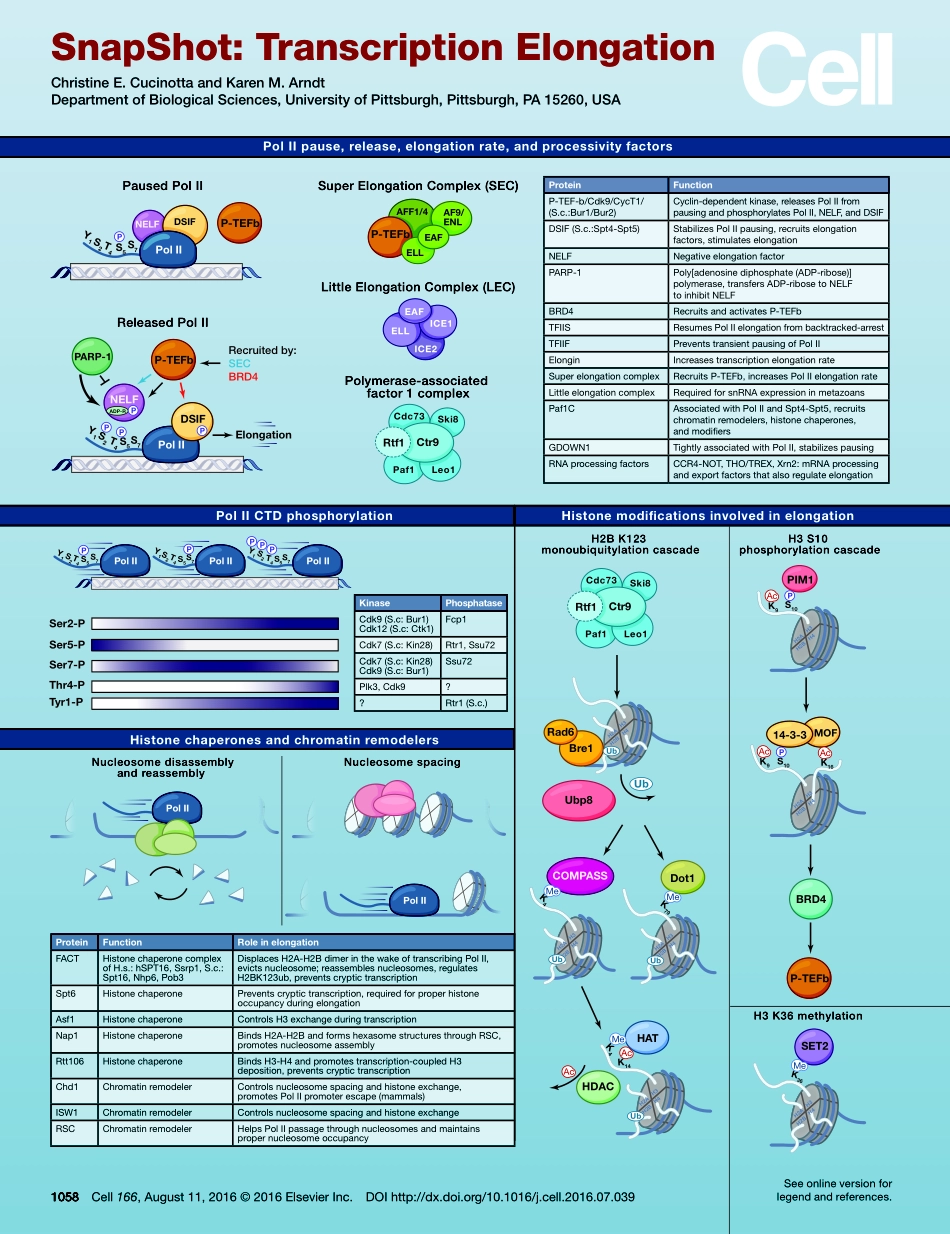
PolIISer5-PSer7-PThr4-PTyr1-PSer2-PY1S2T4S5S7Y1S2T4S5S7Y1S2T4S5S7PolIIpause,release,elongationrate,andprocessivityfactorsK4K79K4K14K9S10Dot1K9S10K16K36PausedPolIIPolIIPolIIPolIICdc73Ski8Rtf1Ctr9Paf1Leo1Cdc73Ski8Rtf1Ctr9Paf1Leo1Rad6Bre114-3-3MOFBRD4PIM1SET2HATHDACCOMPASSSuperElongationComplex(SEC)LittleElongationComplex(LEC)Polymerase-associatedfactor1complexReleasedPolIINucleosomedisassemblyandreassemblyPolIICTDphosphorylationHistonechaperonesandchromatinremodelersNucleosomespacingHistonemodificationsinvolvedinelongationH2BK123monoubiquitylationcascadeH3S10phosphorylationcascadeH3K36methylationAF9/ENLAFF1/4P-TEFbELLEAFICE2EAFICE1ELLPolIIPolIIPolIIPolIIY1S2T4S5S7PolIIP-TEFbDSIFP-TEFbNELFY1S2T4S5S7ADP-RPolIIRecruitedby:SECBRD4DSIFNELFElongationP-TEFbPARP-1PPPPPPPPUbUbUbPPPPMeMeMeMeAcAcAcAcAcH2AH2BH3H4H2AH2BH3H4H2AH2BH3H4H2AH2BH3H4H2AH2BH3H4H2AH2BH3H4H2AH2BH3H4UbUbUbp81058Cell166,August11,2016©2016ElsevierInc.DOIhttp://dx.doi.org/10.1016/j.cell.2016.07.039Seeonlineversionforlegendandreferences.SnapShot:TranscriptionElongationChristineE.CucinottaandKarenM.ArndtDepartmentofBiologicalSciences,UniversityofPittsburgh,Pittsburgh,PA15260,USAProteinFunctionP-TEF-b/Cdk9/CycT1/(S.c.:Bur1/Bur2)Cyclin-dependentkinase,releasesPolIIfrompausingandphosphorylatesPolII,NELF,andDSIFDSIF(S.c.:Spt4-Spt5)StabilizesPolIIpausing,recruitselongationfactors,stimulateselongationNELFNegativeelongationfactorPARP-1Poly[adenosinediphosphate(ADP-ribose)]polymerase,transfersADP-ribosetoNELFtoinhibitNELFBRD4RecruitsandactivatesP-TEFbTFIISResumesPolIIelongationfrombacktracked-arrestTFIIFPreventstransientpausingofPolIIElonginIncreasestranscriptionelongationrateSuperelongationcomplexRecruitsP-TEFb,increasesPolIIelongationrateLittleelongationcomplexRequiredforsnRNAexpressioninmetazoansPaf1CAssociatedwithPolIIandSpt4-Spt5,recruitschromatinremodelers,histonechaperones,andmodifiersGDOWN1TightlyassociatedwithPolII,stabilizespausingRNAprocessingfactorsCCR4-NOT,THO/TREX,Xrn2:mRNAprocessingandexportfactorsthat...