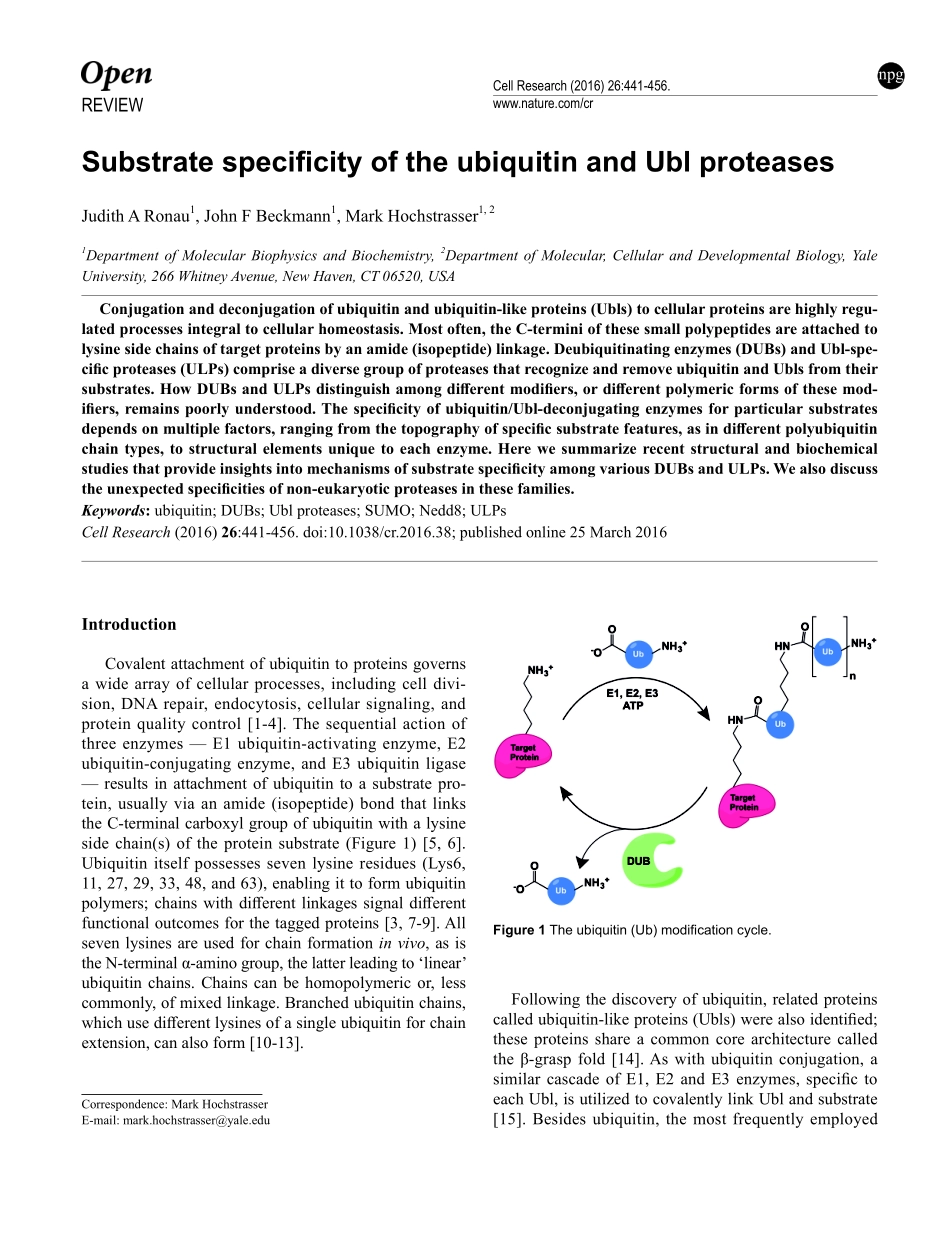
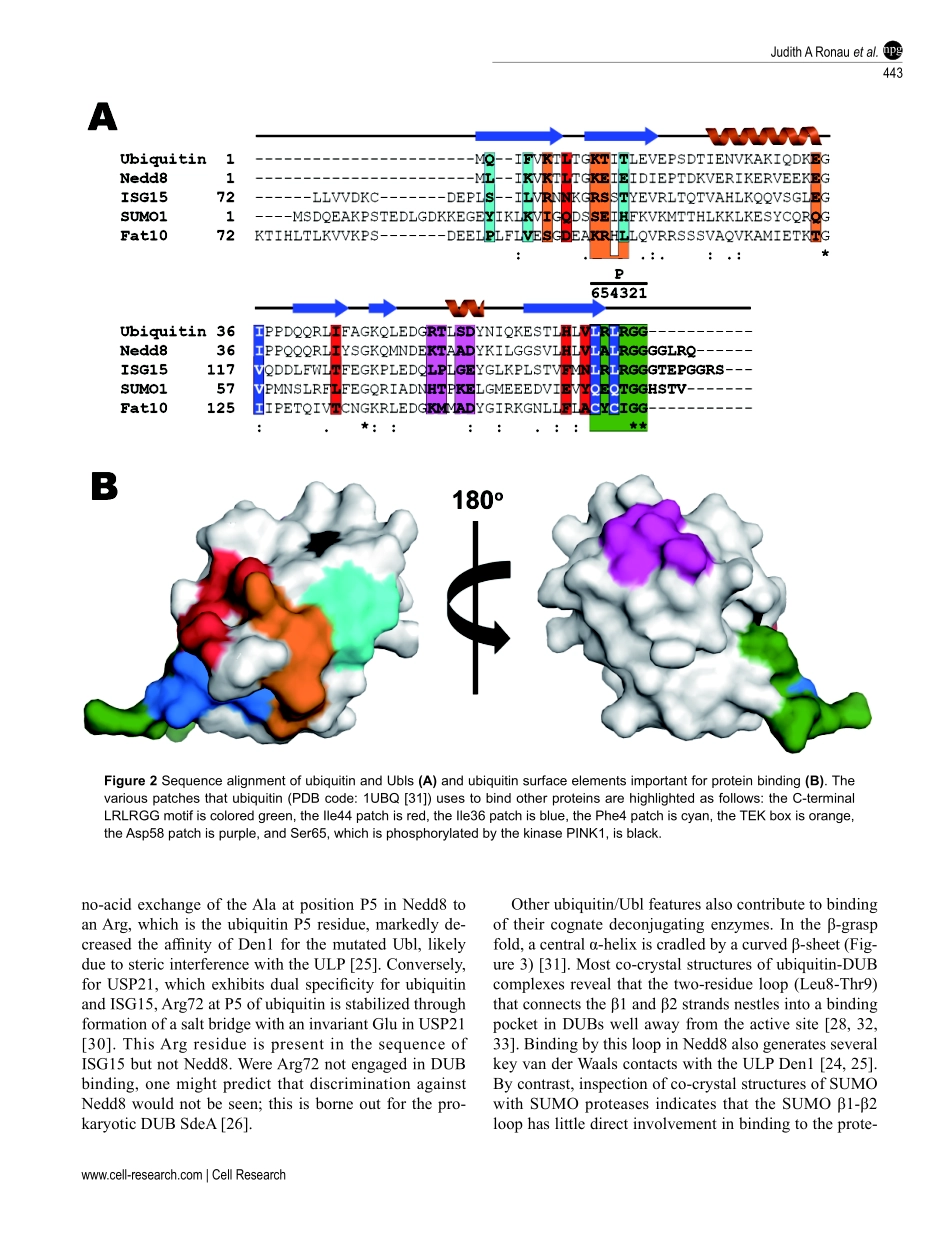
REVIEWnpgCellResearch(2016)26:441-456.www.nature.com/crSubstratespecificityoftheubiquitinandUblproteasesJudithARonau1,JohnFBeckmann1,MarkHochstrasser1,21DepartmentofMolecularBiophysicsandBiochemistry,2DepartmentofMolecular,CellularandDevelopmentalBiology,YaleUniversity,266WhitneyAvenue,NewHaven,CT06520,USAConjugationanddeconjugationofubiquitinandubiquitin-likeproteins(Ubls)tocellularproteinsarehighlyregu-latedprocessesintegraltocellularhomeostasis.Mostoften,theC-terminiofthesesmallpolypeptidesareattachedtolysinesidechainsoftargetproteinsbyanamide(isopeptide)linkage.Deubiquitinatingenzymes(DUBs)andUbl-spe-cificproteases(ULPs)compriseadiversegroupofproteasesthatrecognizeandremoveubiquitinandUblsfromtheirsubstrates.HowDUBsandULPsdistinguishamongdifferentmodifiers,ordifferentpolymericformsofthesemod-ifiers,remainspoorlyunderstood.Thespecificityofubiquitin/Ubl-deconjugatingenzymesforparticularsubstratesdependsonmultiplefactors,rangingfromthetopographyofspecificsubstratefeatures,asindifferentpolyubiquitinchaintypes,tostructuralelementsuniquetoeachenzyme.HerewesummarizerecentstructuralandbiochemicalstudiesthatprovideinsightsintomechanismsofsubstratespecificityamongvariousDUBsandULPs.Wealsodiscusstheunexpectedspecificitiesofnon-eukaryoticproteasesinthesefamilies.Keywords:ubiquitin;DUBs;Ublproteases;SUMO;Nedd8;ULPsCellResearch(2016)26:441-456.doi:10.1038/cr.2016.38;publishedonline25March2016Correspondence:MarkHochstrasserE-mail:mark.hochstrasser@yale.eduIntroductionCovalentattachmentofubiquitintoproteinsgovernsawidearrayofcellularprocesses,includingcelldivi-sion,DNArepair,endocytosis,cellularsignaling,andproteinqualitycontrol[1-4].Thesequentialactionofthreeenzymes—E1ubiquitin-activatingenzyme,E2ubiquitin-conjugatingenzyme,andE3ubiquitinligase—resultsinattachmentofubiquitintoasubstratepro-tein,usuallyviaanamide(isopeptide)bondthatlinkstheC-terminalcarboxylgroupofubiquitinwithalysinesidechain(s)oftheproteinsubstrate(Figure1)[5,6].Ubiquitinitselfpossessessevenlysineresidue...