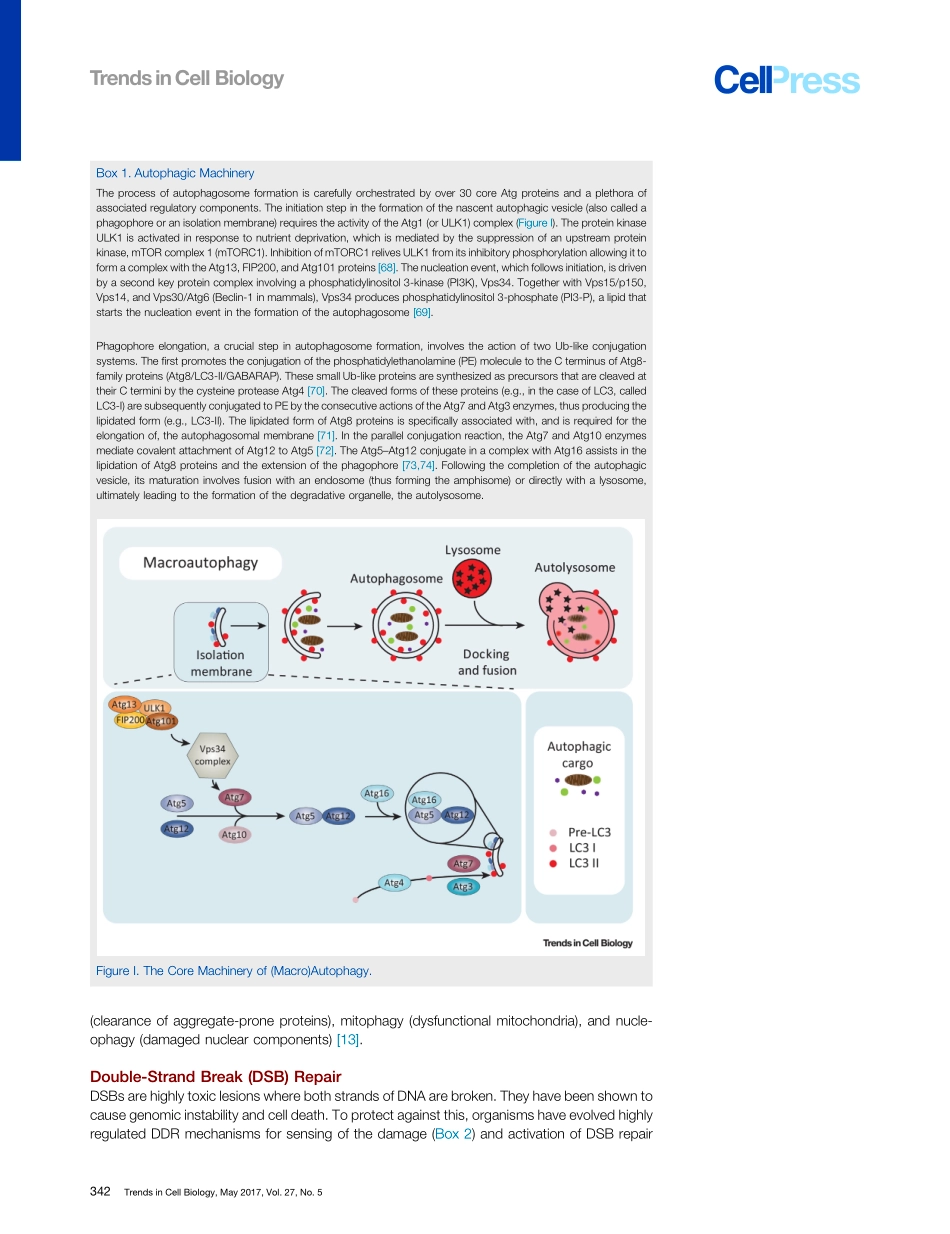
ReviewRepair,Reuse,Recycle:TheExpandingRoleofAutophagyinGenomeMaintenanceGraemeHewitt1,*andViktorI.Korolchuk2,*(Macro)Autophagyisacatabolicpathwaythatdeliversexcess,aggregated,ordamagedproteinsandorganellestolysosomesfordegradation.Autophagyisactivatedinresponsetonumerouscellularstressorssuchasincreasedlevelsofreactiveoxygenspecies(ROS)andlowlevelsofcellularnutrientsaswellasDNAdamage.Althoughautophagyoccursinthecytoplasm,itsinhibitionleadstoaccumulationofDNAdamageandgenomicinstability.Inthepastfewyears,ourunderstandingoftheinterplaybetweenautophagyandgenomicstabilityhasgreatlyincreased.Inthisreviewwesummarizetheserecentadvancesinunder-standingthemolecularmechanismslinkingautophagytoDNArepair.IntroductionMaintenanceofgenomicintegrityisessentialfororganismalsurvival.DNAcanbedamagedbyaplethoraofextrinsicfactorssuchasUVradiation,ionizingradiation,andchemicalcompoundsaswellasintrinsicfactorssuchasfreeradicalsgeneratedaspartofnormalmetabolismandmistakesinreplication.Thisbarrageofgenotoxicinsultsresultsinthegenerationofanestimated19200DNAlesionsperday[1].Itisthereforenecessaryforcellstohaveeffectivemechanismsforsensingandrepairingthisdamagetomaintaintheirsurvival.Likegenomicintegrity,maintenanceofproteinhomeostasisisanessentialrequirementforcellandorganismalsurvival.Thus,therearemultiplepathwaysresponsiblefortheturnoverofdamagedandunwantedproteinsandorganelles.Theubiquitin(Ub)proteasomesystem(UPS)isresponsibleforthedegradationofshort-lived,solubleproteins,isactiveinboththecytoplasmandthenucleus,andisthoughttoaccountforthemajorityofproteinturnoverinthecell.TaggingoftargetproteinswithUbchainsmakestheUPShighlyselective.Asaresultofitsabilitytotightlycontrolproteinlevels,theUPShasbeenshowntoregulatenumerouscellularprocessessuchasthecellcycle,signaltransduction,andDNArepair[2].Theterm‘autophagy’translatesfromtheGreekautomeaning‘oneself’andphagymeaning‘toeat’anddescribesevolutionarilyconservedcataboliccellulardegradationpathwaysinvolvedinthedeliveryofcytoplasmiccargototh...