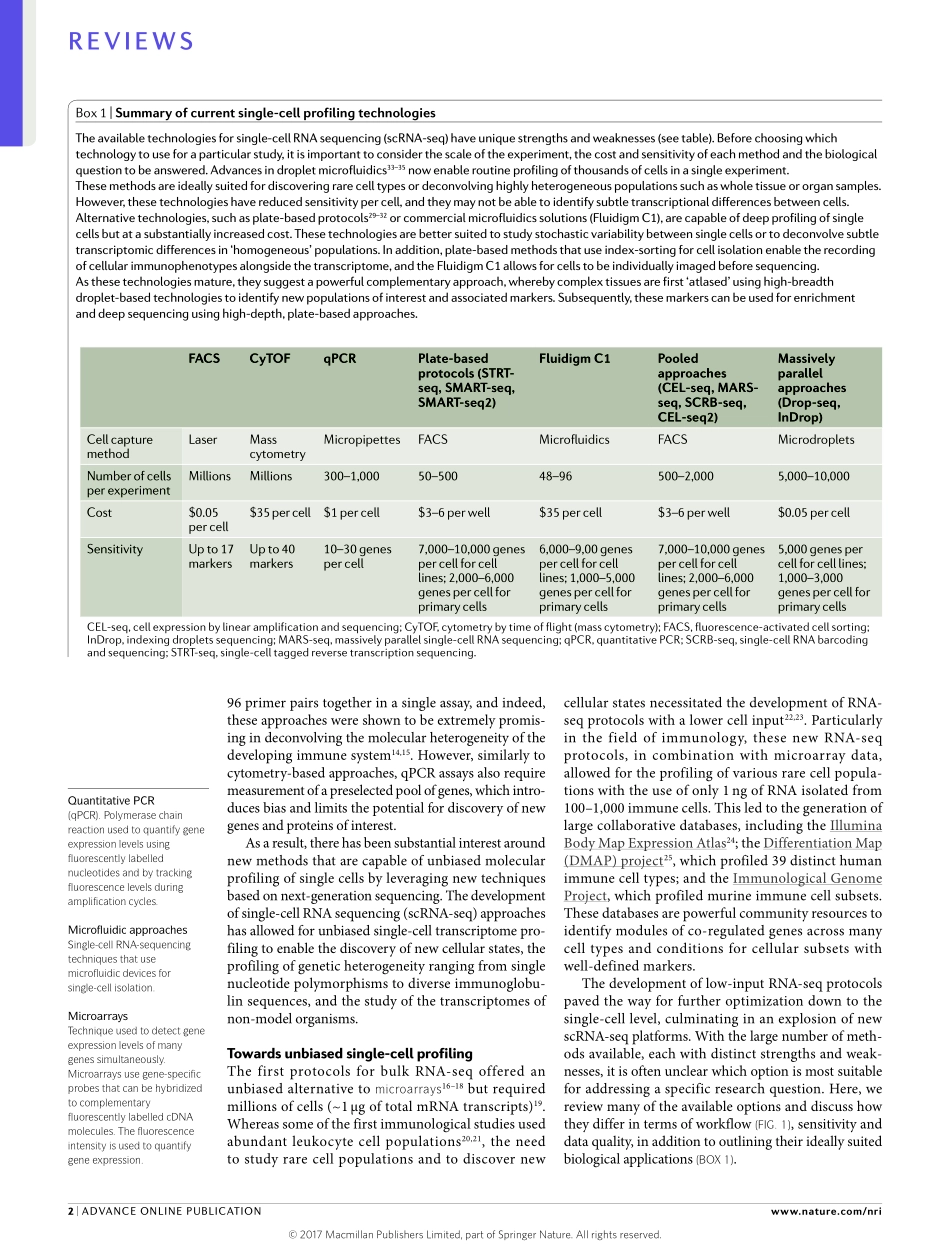
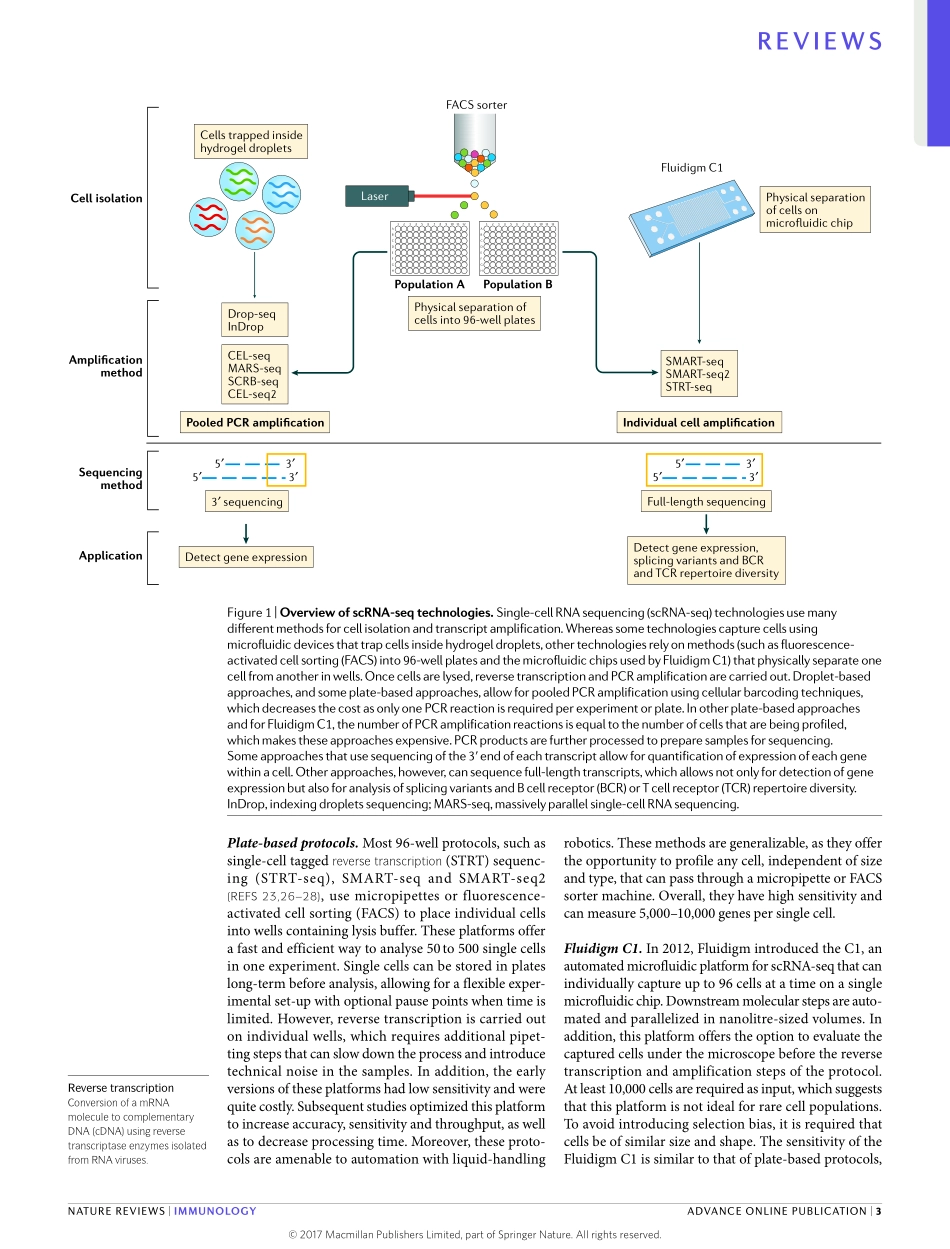
Frombacteriatohumans,thediverseandadaptablenatureofforeignthreatshasdriventheevolutionofapowerfulandflexibledefenceresponse.Tomaintainitseffectiveness,thisso‑calledimmunesystemhaspro‑ducedhighlyspecialized(pathogen‑specific)celltypesthatworktogethertoprevent,retainamemoryofandeliminatedisease1–4.Single‑cellresolutionisthereforeessentialtounderstandinghowtheimmunesystemgivesrisetosuchabreadthofpotentialresponsesagainstmanydifferentpathogens5.Recently,newtechnologieshavebeendevelopedthatenabletheprofilingofsinglecellsusingnext‑generationsequencing,whichoffersanunbiasedapproachtostudyingimmunecelldiver‑sity.InthisReview,wepresentanoverviewofexistingsingle‑celltechnologiesanddiscusstheirstrengthsandlimitations(BOX1).Wealsoexplorewaysinwhichtheseapproachescandeepenourunderstandingofimmunologicalresponsesanddisease,andweexaminecutting‑edgetrendsandpotentialfutureinnovationsinthefield.‘Targeted’single-cellprofilingtechnologiesAlargenumberoftechniqueshaveleveragedadvancesinmicroscopy,cytometry,molecularbiologyand,mostrecently,next‑generationsequencingtoprofilesinglecells.Manyoftheseapproacheshavebeendevelopedandoptimizedtobeusedinstudiesthataimtodecon‑volveimmunecellheterogeneity,buttheycandifferbyordersofmagnitudeintermsofthenumberofcellsthatcanbeanalysedperexperiment(thebreadthofcellularprofiling)andthenumberofgenespercellthatcanbedetected(thedepthofcellularprofiling).‘Targeted’technologiescanassessapre‑selectedsetofmoleculardimensions(pre‑selectedgenesformRNAexpressionstudiesandprotein‑leveldetection)acrosshundredstomillionsofcellsusingknownmolecularbaits—suchasfluorescentlylabelledoligonucleotideprobes,fluorescentormetal‑conjugatedantibodies,orPCRprimers—toprofilegenesorproteinswithsingle‑cellresolution.Forexample,recentadvancesinflowcytometry6haveallowedfortheroutineandsimul‑taneousprofilingofupto17proteinspercellusingfluorescentantibodies.Byusingmetal‑conjugatedanti‑bodiestoovercomethespectrallimitsoffluorescentproteins,masscytometry7canfur...