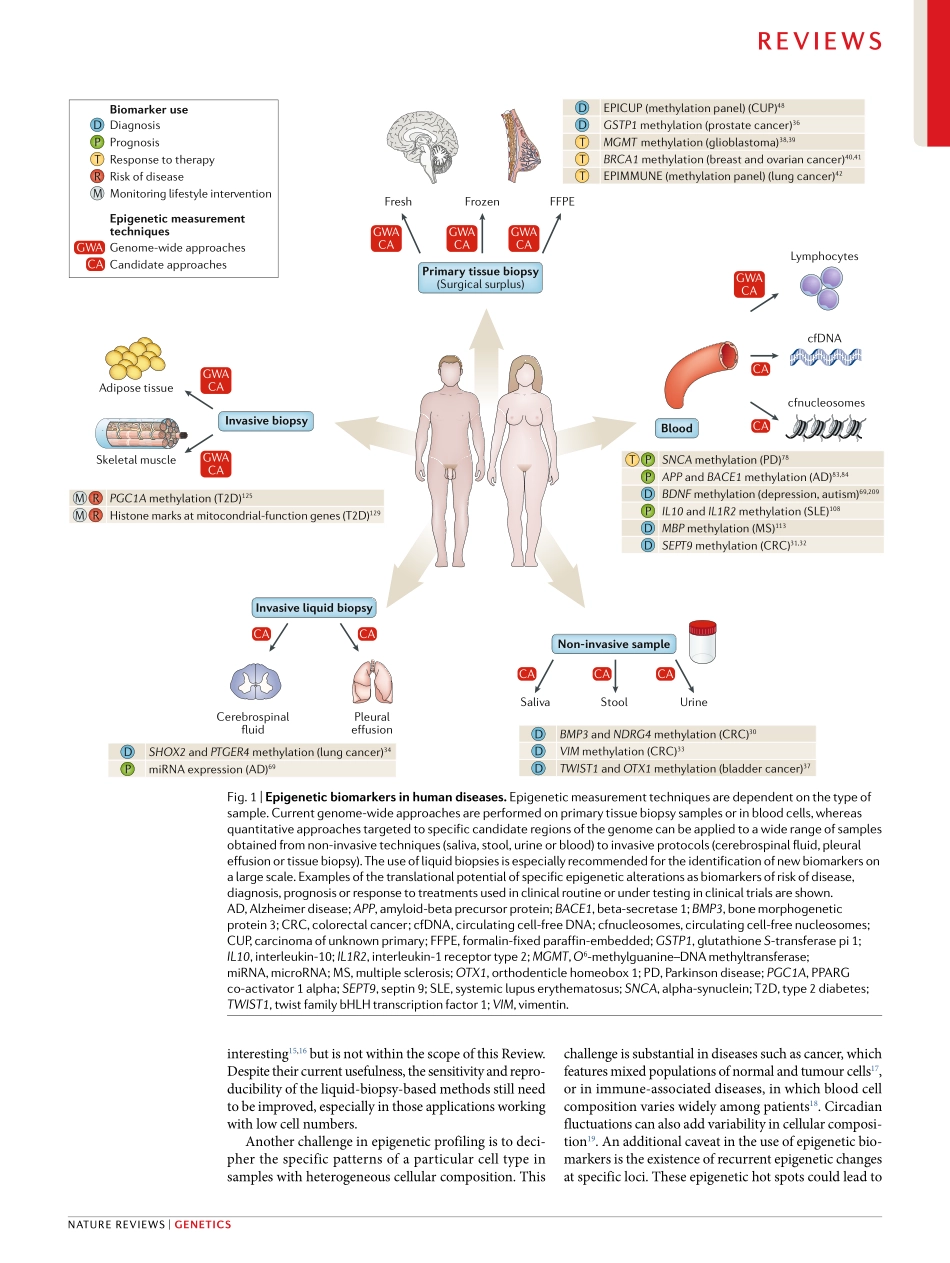
Forthepasttwodecades,thestudyofgeneticvariantshasheldthemostpromiseinpersonalizedmedicine.TheHumanGenomeProjectwasattheforefrontofseveralinternationaleffortstofullydescribethegenomeinbothnormalanddiseasedtissue1.Itisindisputablethattheknowledgegleanedfromthesegeneticstudieshasstronglycontributedtoourunderstandingofthemolec-ularbasisofseveralhumandisordersandhasevenprovidedafewcasesthataretranslatedintoclinicalpractice.However,throughthislongprocess,wehavelearnedthatthephenotypeofcomplexdiseasestatescannotbefullyexplainedbysinglegeneticvariantsandthattherolesofspecificgenesmayhavebeenoveres-timated2.Forcomplextraitssuchascommondiseases,therisk,progressionandresponsetotreatmentaredif-ficulttomodelandleverageinclinicaldecision-makingastheydonotfollowclassicgeneticheritability.Despitethedevelopmentofcomputationalmethodstoinferriskpredictionmodelsonthebasisoftheinteractionbetweengeneticvariantsandenvironmentalfactors3,inmostcasescommongenetictraitsdonotproduceacon-sistentphenotype.Asthecombinedeffectsofknowntrait-associatedgeneticvariantstypicallyexplainonlypartoftheheritabilityofcomplextraits,theterm‘miss-ingheritability’wascoinedtohighlightthepolygenicnatureandtheresponsetotheenvironmentofcomplexphenotypes4.Epigeneticsprovidesamolecularexplanationtobridgethegapbetweenthegenomeandenvironmentalsignalsduringdevelopmentandcanbeassociatedwithlifestyleandenvironmentalconditionseitherduringintra-uterineorpostnataldevelopment(Box1).Theflexibilityoftheepigenomerepresentsanenticingopportunitytounderstanddiseasevariation,andseveralinitiativestoobtaintheepigenomeofhumandisorderswererapidlydeveloped,particularlyincancer2,5.Despitelimitationsinmethodologiesand/orsamplesize,epigenomicmapsofhumanhealthanddiseasearenowavailableandformacomplexnetworkwithgeneticandenvironmentaldata.InthisReview,weprovideacomprehensivestate-of-the-artoverviewofclinicalepigeneticsinmajorhumandisorders.Oncologyisthemainfocusintrans-lationalepigenetics,withbiomarkersapprovedbytheUSFoodandDrugAdministrat...