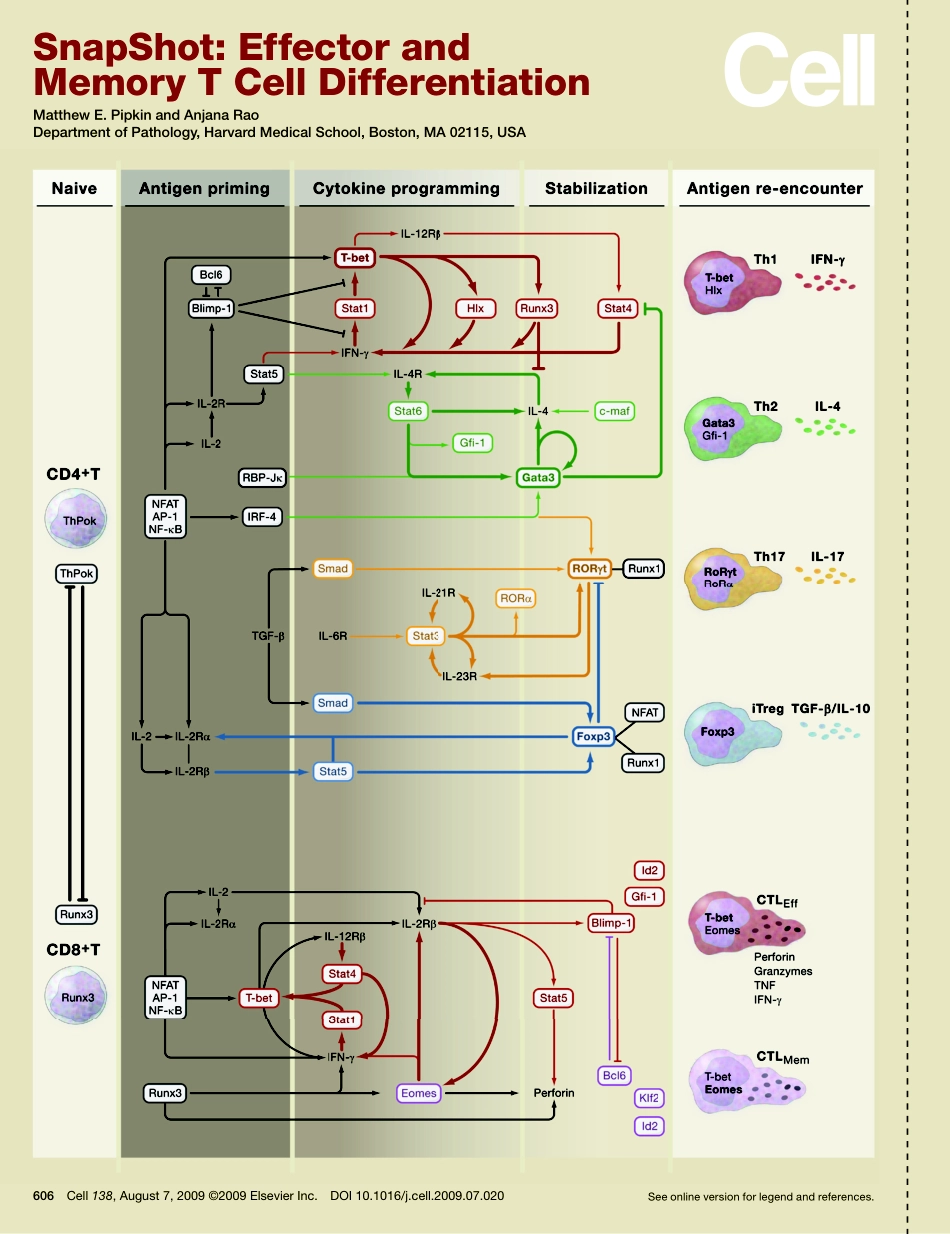
SnapShot:EffectorandMemoryTCellDifferentiationMatthewE.PipkinandAnjanaRaoDepartmentofPathology,HarvardMedicalSchool,Boston,MA02115,USASeeonlineversionforlegendandreferences.606Cell138,August7,2009©2009ElsevierInc.DOI10.1016/j.cell.2009.07.020606.e1Cell138,August7,2009©2009ElsevierInc.DOI10.1016/j.cell.2009.07.020SnapShot:EffectorandMemoryTCellDifferentiationMatthewE.PipkinandAnjanaRaoDepartmentofPathology,HarvardMedicalSchool,Boston,MA02115,USAThedifferentiationofTcellsisanidealsystemtostudythemolecularbasisoflineagespecificationinmammaliancells.Uponstimulationwithantigenduringinfectionorinflammation,naiveperipheralTcellsdifferentiateintovarioustypesofeffectorTcellswithspecificimmunefunctions.NaiveCD4+Tcellsdifferentiateintoatleastfoursubsets(lineages)ofThelper(Th)cells:Th1,Th2,Th17,or“induced”regulatoryTcells(iTregs).Eachsubsetisdistinguishedbythecytokinesthattheyproduce(Anseletal.,2006;Leeetal.,2006;Zhouetal.,2009).NaiveCD8+TcellsdifferentiateintoeffectorcytolyticTlymphocytes(CTLEff)thatkillinfectedhostcellsusingthepore-formingproteinperforinandserineesterasescalledgranzymes(Cruz-Guillotyetal.,2009).Alternatively,naiveCD8+TcellscandifferentiateintomemoryCTLs(CTLMem)thatsurvivelong-termandprotectthehostfromreinfection(KaechandWherry,2007).Tcelldifferentiationisinlargepartdeterminedbysignalsfromtheenvironmentandisshapedbynumerousfeedbackandfeed-forwardloops(boldarrows)thatmodulateandreinforcethedirectioninwhichdifferentiationproceeds(Singh,2007).Transcriptionfactors(boxes)playakeyroleinthisprocessbyformingnetworksinwhichtheyreinforceoropposeeachother’sactions.ThisSnapShotillustratesthedifferentiationpathwaysforseveralofthebestcharacterizedTcellsubsets.NaiveThetranscriptionfactorThPokandtranscriptionfactorsoftheRunxfamilyactinanantagonisticfashiontospecifythedevelopmentofnaiveCD4+andCD8+αβTcellsthatemigratefromthethymusandcolonizeperipherallymphoidorgans(Taniuchi,2009).AntigenprimingUponencounteringantigenandwithcostimulationbyantigen-presentingcells,naiveT...