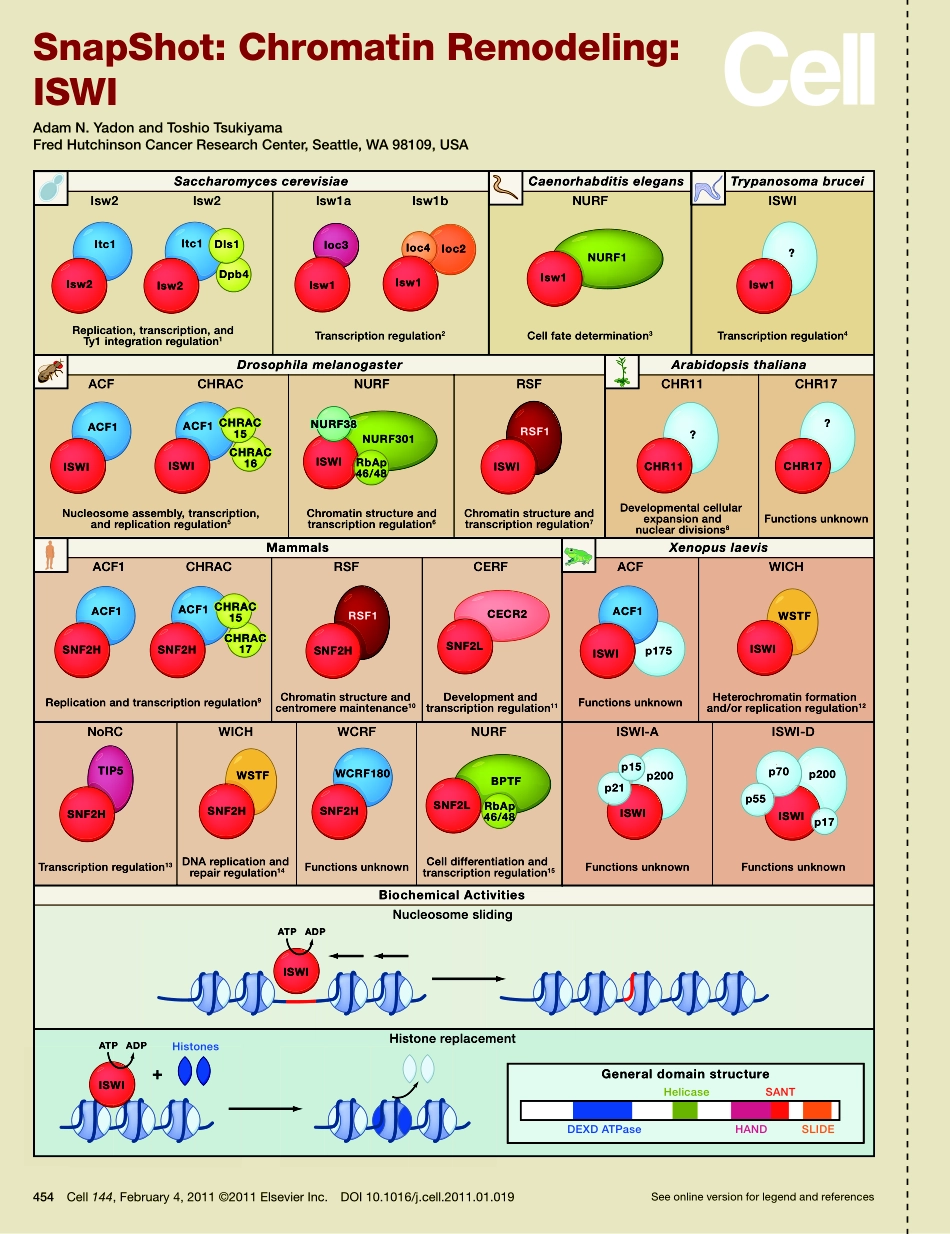
Seeonlineversionforlegendandreferences454Cell144,February4,2011©2011ElsevierInc.DOI10.1016/j.cell.2011.01.019SnapShot:ChromatinRemodeling:ISWIAdamN.YadonandToshioTsukiyamaFredHutchinsonCancerResearchCenter,Seattle,WA98109,USAGeneraldomainstructureDEXDATPaseHANDHelicaseSANTSLIDEIsw2ACFCHRACIsw2Isw1aIsw1bReplication,transcription,andTy1integrationregulation1Nucleosomeassembly,transcription,andreplicationregulation5Chromatinstructureandtranscriptionregulation6Chromatinstructureandtranscriptionregulation7Replicationandtranscriptionregulation9Transcriptionregulation13DNAreplicationandrepairregulation14FunctionsunknownCelldifferentiationandtranscriptionregulation15FunctionsunknownFunctionsunknownChromatinstructureandcentromeremaintenance10Developmentandtranscriptionregulation11CERFFunctionsunknownHeterochromatinformationand/orreplicationregulation12Developmentalcellularexpansionandnucleardivisions8FunctionsunknownTranscriptionregulation2Cellfatedetermination3Transcriptionregulation4Itc1Isw2NoRCWICHNURFNucleosomeslidingHistonereplacementISWI-AISWI-DWCRFACF1SNF2HWCRF180SNF2HACFACF1WICHRSFp175ACF1ISWIWSTFISWIISWIRSF1Isw1Ioc3NURFISWINURFRSFCHR11CHR17CHRACSaccharomycescerevisiaeCaenorhabditiselegansDrosophilamelanogasterArabidopsisthalianaXenopuslaevisMammalsBiochemicalActivitiesTrypanosomabruceiItc1Isw2Dpb4Dls1Ioc2Ioc4Isw1NURF1Isw1?Isw1ACF1ISWIACF1ISWI?CHR11?CHR17NURF301ISWIACF1SNF2HRSF1SNF2HSNF2LCECR2ATPADPTIP5SNF2HWSTFSNF2HSNF2LBPTFRbAp46/48RbAp46/48ISWIp200p15p21p70p200p17ISWIp55ISWIISWIATPADP+HistonesNURF38NURF38CHRAC15CHRAC15CHRAC15CHRAC15CHRAC17CHRAC17CHRAC16CHRAC16454.e1Cell144,February4,2011©2011ElsevierInc.DOI10.1016/j.cell.2011.01.019SnapShot:ChromatinRemodeling:ISWIAdamN.YadonandToshioTsukiyamaFredHutchinsonCancerResearchCenter,Seattle,WA98109,USATheimitationswitch(ISWI)familyofATP-dependentchromatin-remodelingenzymescompriseshighlyconservedproteincomplexesthatutilizetheenergyofATPhydrolysistoslidenucleosomesalongDNAand/orreplacehistoneswithinnucleosomes.AllAT...