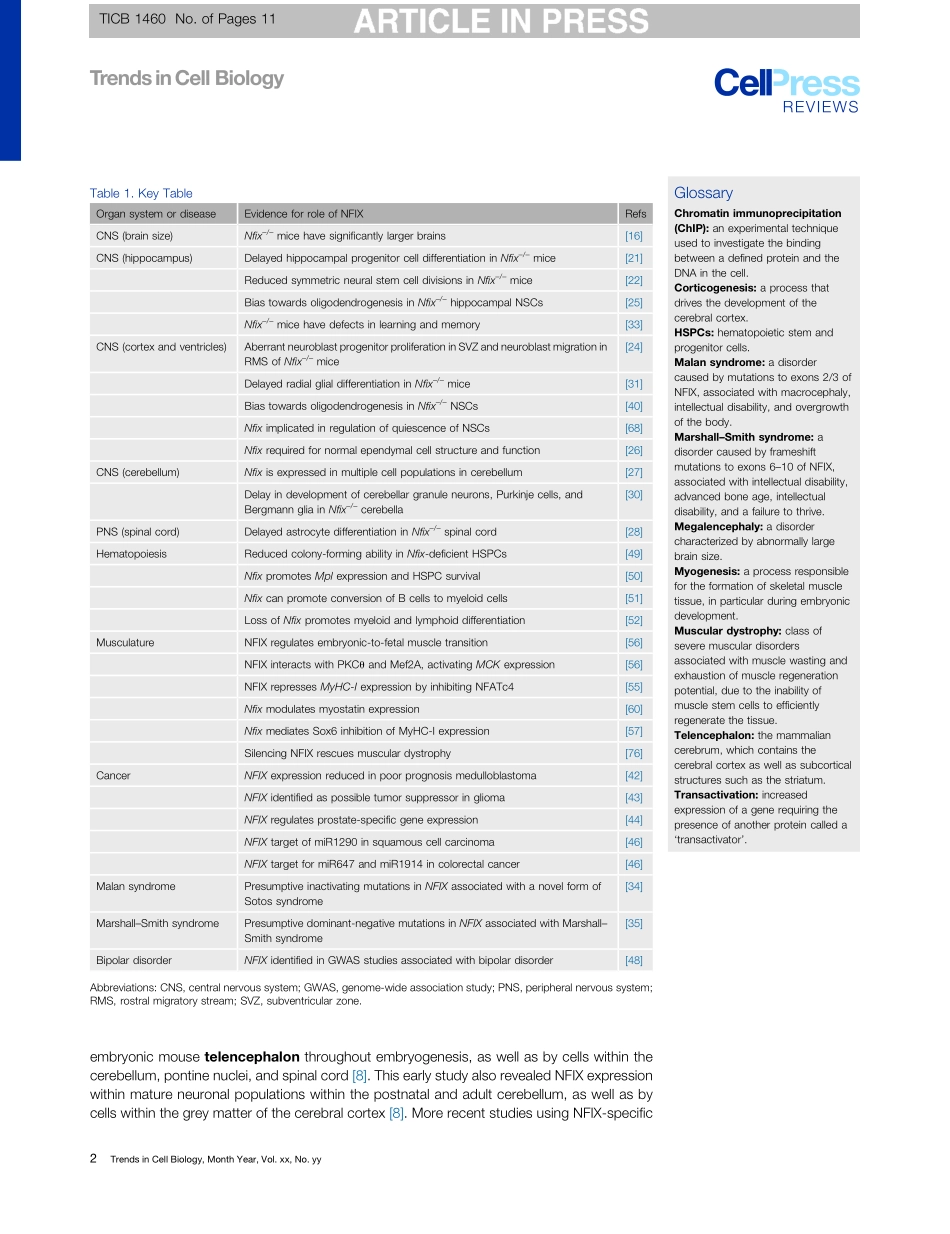
ReviewNuclearFactorOneXinDevelopmentandDiseaseMichaelPiper,1,*RichardGronostajski,2andGraziellaMessina3,*Thepastdecadehasseenincredibleadvancesinthefieldofstemcellbiologythathavegreatlyimprovedourunderstandingofdevelopmentandprovidedimportantinsightsintopathologicalprocesses.Transcriptionfactors(TFs)playacentralroleinmediatingstemcellproliferation,quiescence,anddifferentiation.OneTFthatcontributestotheseprocessesisNuclearFactorOneX(NFIX).Recently,NFIXactivityhasbeenshowntobeessentialinmultipleorgansystemsandtohaveimportanttranslationalimpactsforhumanhealth.Here,wedescriberecentstud-iesshowingthecontributionofNFIXtomuscledevelopmentandmusculardystrophies,hematopoiesis,cancer,andneuralstemcellbiology,highlightingtheimportanceofthisknowledgeinthedevelopmentoftherapeutictargets.NFIXandtheNFIFamilyNFIXisamemberoftheNFI(nuclearfactorI)TFfamilythatalsoincludesNFIA,NFIB,andNFIC.TheNFIfamilywasdiscoveredasaproteinfractionfromhumancellsthatwasessentialfortheinvitroreplicationofadenovirustype4(Ad4)DNA[1]andwasthenshowntopossesssite-specificDNA-bindingactivityspecificforbothAd4replicationorigins[2]andcellularsites[3].TwosubsequentstudiesindependentlydiscoveredtheNFIproteins,namingthemasTGGCA-bindingprotein[4]andCAAT-boxTF[5].GenesencodingallfourNFIfamilymemberswerefirstclonedfromchicken[6,7]andthenfrommouse[8];indeed,fourhomologousgenesexistinallmammals[9].AllfourgenesencodeproteinswithahighlyconservedN-terminalDNA-bindinganddimerizationdomain.TheC-terminaltransactivation/repression(seeGlossary)domainsofeachNFIfamilymemberexhibitmarkedlylowerconservationbetweenfamilymembers[5].mRNAfromNFIgenescanbealternativelyspliced,yieldinguptoadozenisoforms[10].OnlysingleNFIgenesarepresentinnon-vertebrates,includingCaenorhabditiselegansandDro-sophila[9,11].NFIproteinsbindashomo-orhetero-dimers[12]tothedyadconsensussequenceTTGGCN5GCCAAandbindspecifically,butlesstightly,to1/2sites(i.e.,TTGGCorGCCAA)[13,14].DifferentialexpressionofNfixduringmouseembryogenesiswasanearlyindicationthatthisgene...