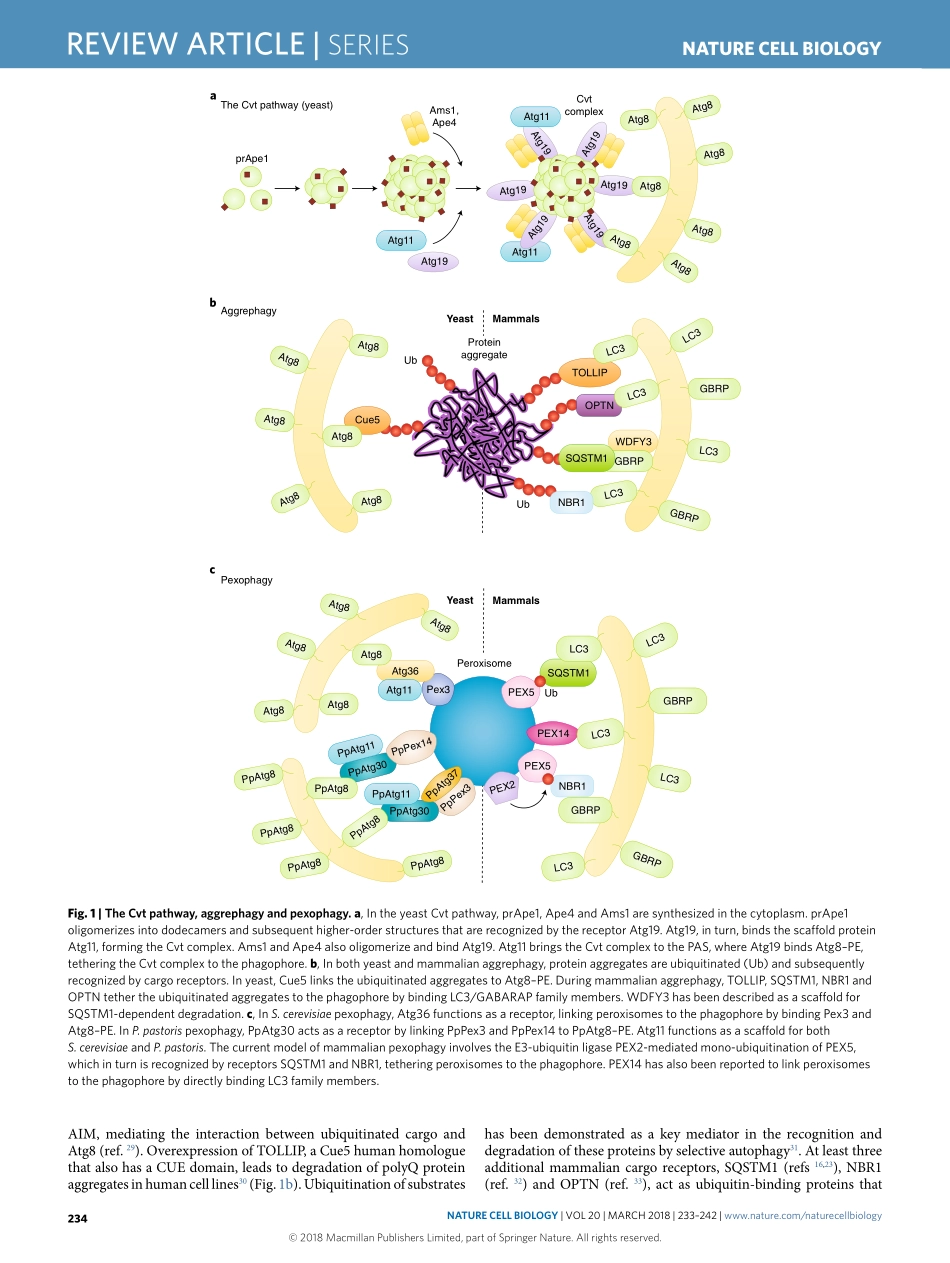
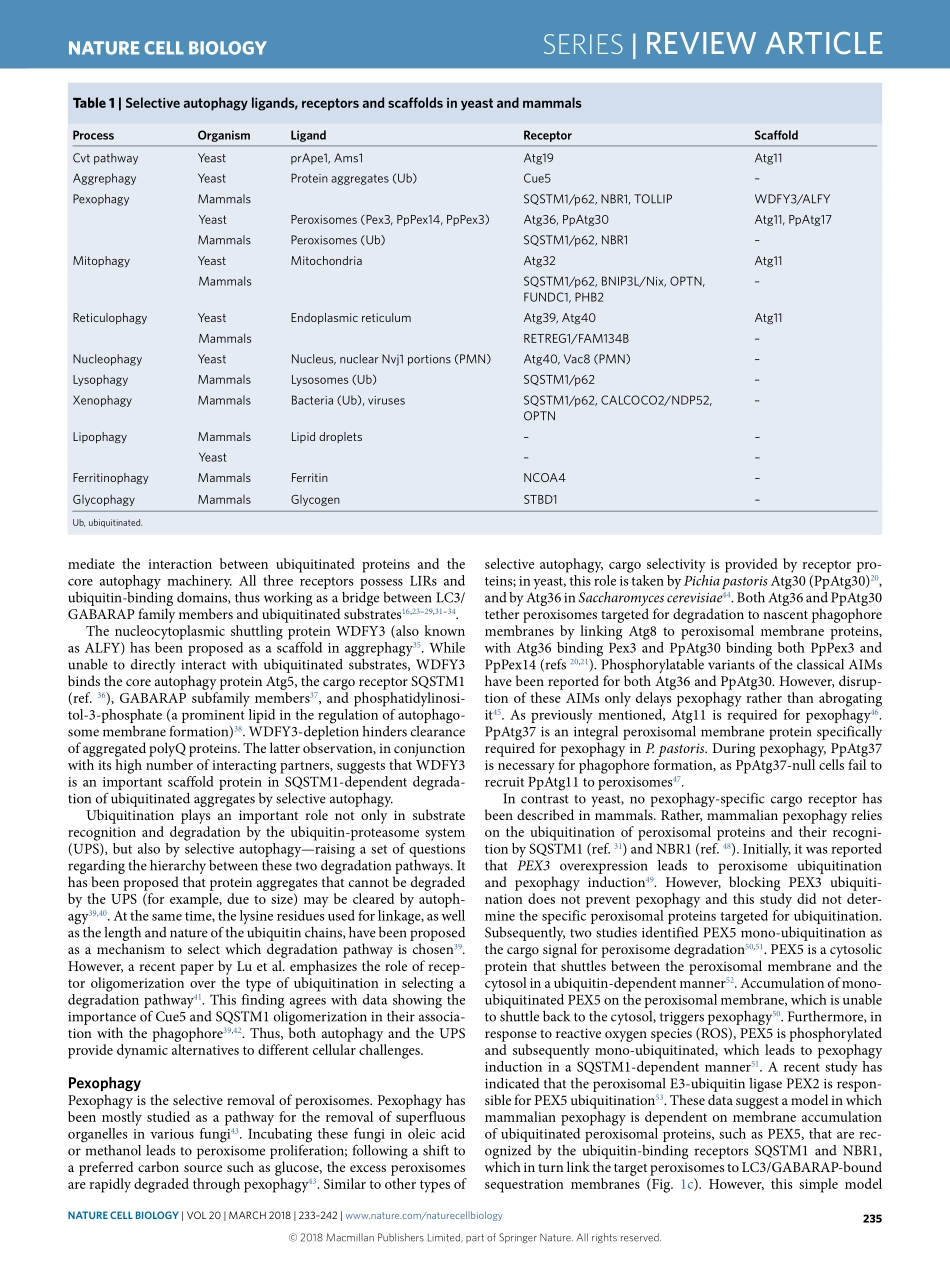
SerieS|REVIEWARTICLEhttps://doi.org/10.1038/s41556-018-0037-zLifeSciencesInstituteandDepartmentofMolecular,CellularandDevelopmentalBiology,UniversityofMichigan,AnnArbor,MI,USA.*e-mail:Klionsky@umich.eduAutophagyisahighlyconservedpathwayineukaryotes,involv-ingcellularrecyclingofmultiplecytoplasmiccomponentsduringstandardphysiologicalconditionsandinresponsetodifferenttypesofstress,suchasstarvation.Macroautophagy(hereafterautophagy)canbeeithernon-selectiveorselectiveandinvolvesthesequestrationofcytoplasmwithindouble-membranevesiclestermedautophagosomes.Uponmaturation,autophago-somesfusewiththevacuole(inyeastandplants)orendosomesandlysosomes(inmetazoans),leadingtodegradationoftheircargobyresidenthydrolases1,2.Cargodegradationproducesmolecularbuild-ingblockssuchasaminoacids,whicharesubsequentlyrecycledbackintothecytoplasmforreuse1,3.Whereasnon-selectiveautoph-agy,acellularresponsetonutrientdeprivation,typicallyinvolvesrandomuptakeofcytoplasmintophagophores(theprecursorstoautophagosomes),selectiveautophagyisresponsibleforspecifi-callyremovingcertaincomponentssuchasproteinaggregatesanddamagedorsuperfluousorganelles4.Differentstudieshavereportedtheselectiveautophagicdegradationofseveralorganelles,includ-ingmitochondria5,peroxisomes6,lysosomes7,endoplasmicreticu-lum(ER)andthenucleus8,undervariousconditions.Furthermore,autophagyselectivelydegradesaggregation-pronemisfoldedpro-teinsandproteinmicroaggregatesimplicatedinthepathologyofvariousneurodegenerativediseases9.InthisReviewArticle,weaddresstheprincipalmechanismsofselectiveautophagyinyeastandmammals,withanemphasisonmitophagy,whichisthebest-describedtypeofselectiveautophagytodate.Cytoplasm-to-vacuoletargetingpathwayThecytoplasm-to-vacuoletargeting(Cvt)pathwayisabiosyn-theticautophagy-relatedprocessspecifictoyeast,inwhichvacu-olarenzymesaretransportedfromthecytoplasmintothevacuoleutilizingtheautophagicmachinery.AmongtheenzymesthatutilizetheCvtpathwayareaminopeptidaseI(Ape1),Ape4andα-man-nosidase(Ams1)10.Ape1isfirstsynthes...