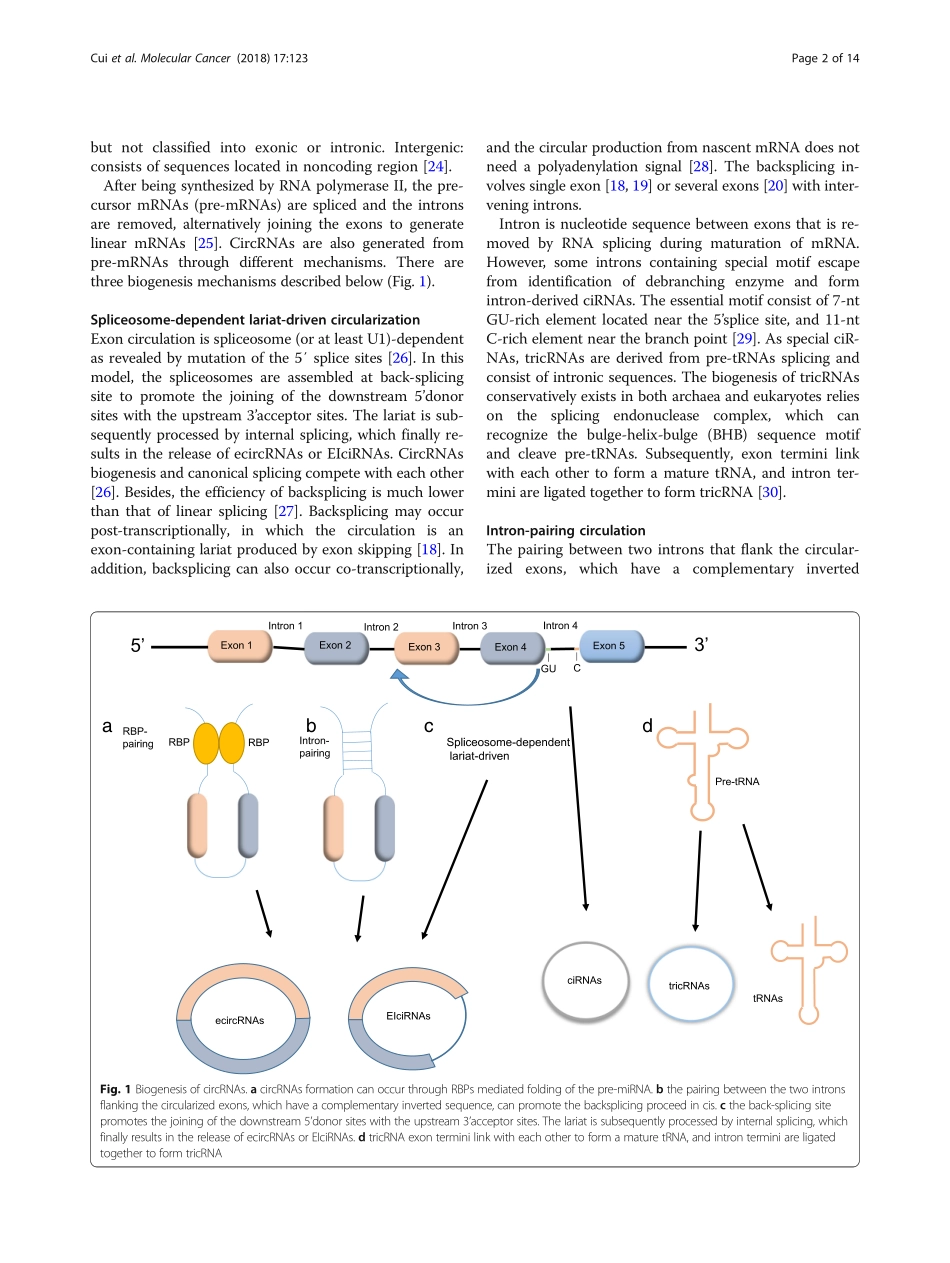
REVIEWOpenAccessEmergingfunctionandpotentialdiagnosticvalueofcircularRNAsincancerXianglunCui1,JianxunWang2,ZongjunGuo3,MengyangLi2,MingyuLi1,SiLiu1,HaoranLiu1,WenjingLi1,XunhuaYin1,JiapingTao1andWenhuaXu1*AbstractAsanovelclassofendogenousRNAs,circRNAs,haveacovalentlyclosedcontinuousloop,withneithera5’to3’polarity,norapolyadenylatedtail.NumerouscircRNAshavebeencharacterizedbyabundance,stabilization,conservation,andexhibittissue/developmentalstage-specificexpression.Furthermore,circRNAsplayvitalrolesintumorigenesisandmetastasis,suchasfunctioningasaceRNAormiRNAsponge,interactingwithproteinandencodingprotein.Increasingevidencehasrevealedthatitpotentiallyservesasarequirednovelbiomarkerforcancerdiagnosis.ThisreviewsummarizedthelatestresearchoncircRNAs,includingitsclassificationandbiogenesis,mechanismandfunctions,aswellascircRNAsindifferentcancers,asapotentialbiomarker.Keywords:CircularRNA,Cancer,Biogenesis,Function,BiomarkerBackgroundAsanovelclassoflongnon-codingRNAs,circularRNAs(circRNAs)arewidelyexpressedinthetreeoflife[1–3].circRNAshaveoriginallybeenconsideredasnon-functionalaccidentalby-productsofaberrantspli-cing[4],whichhasnotreceivedenoughattention.Withtheemergenceofnext-generationsequencing,especiallyRNAsequencingtechnology,numerouscircRNAshavebeenfoundtobeextensivelyexpressedineukaryoticcells.circRNAsaresingle-strandedtranscriptsderivedfromexons,introns,orintergenicregionsthathaveacovalentlyclosedcontinuousloopwithoutapolyadeny-latedtail[5].Duetotheclosedstructure,circRNAshavebeenshowntobehighlystable.NumerouscircRNAsdisplayevolutionaryconservation,andtheexpressionprofilesarecelltype-ordevelopmentalstage-specific.Cancerisoneofthemostseriousandlife-threateningdiseases,whichhashighmorbidityandmortalityworld-wide,andahighfrequencyofmetastasisandrecurrence.Hence,thereisanurgentneedtoidentifypotentialbio-markersforprognosispredication,anddeterminenewtargetstodesignmorepowerfultherapeuticapproaches.VariousstudieshavesuggestedthatcircRNAsareofgreatsignificanceintu...