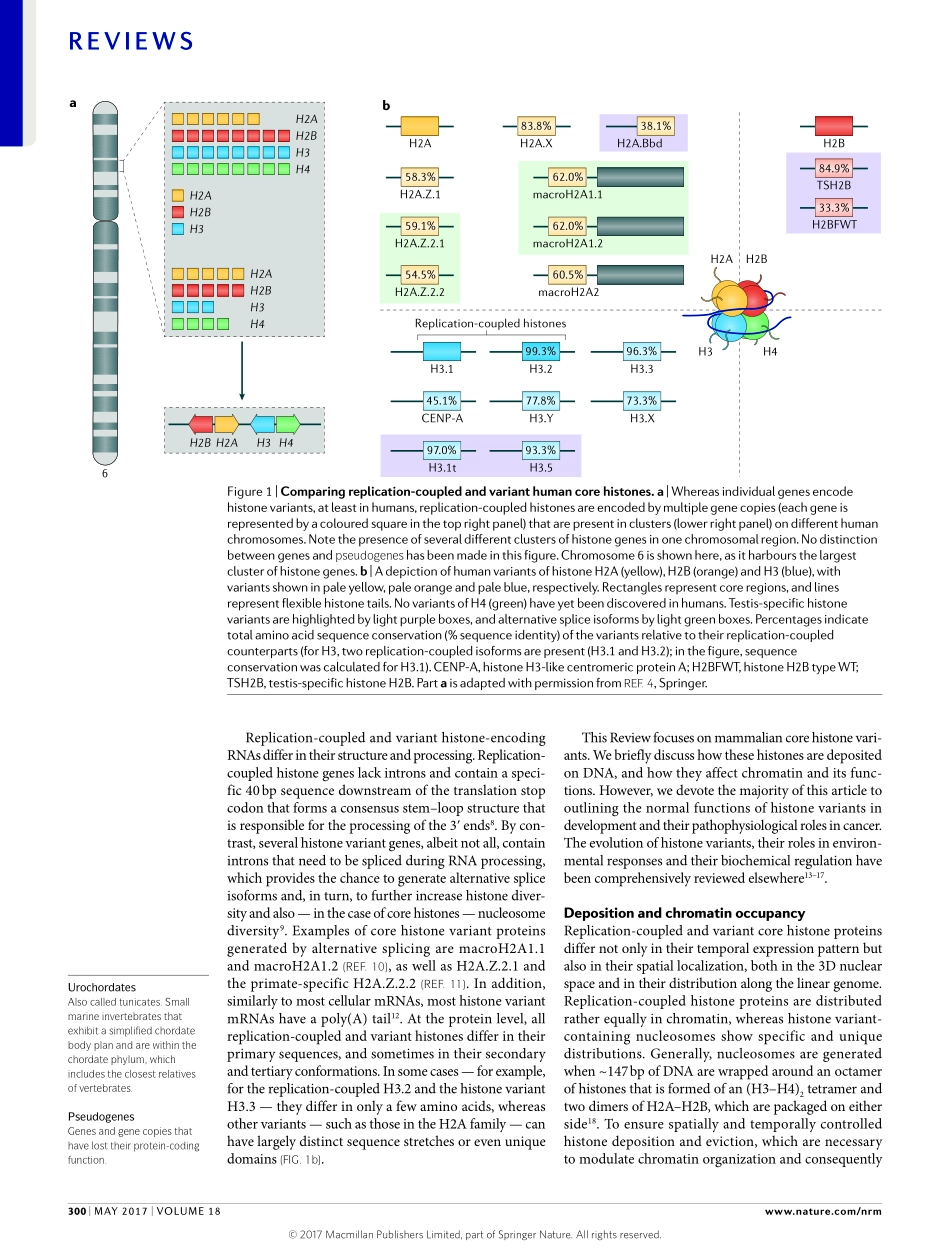
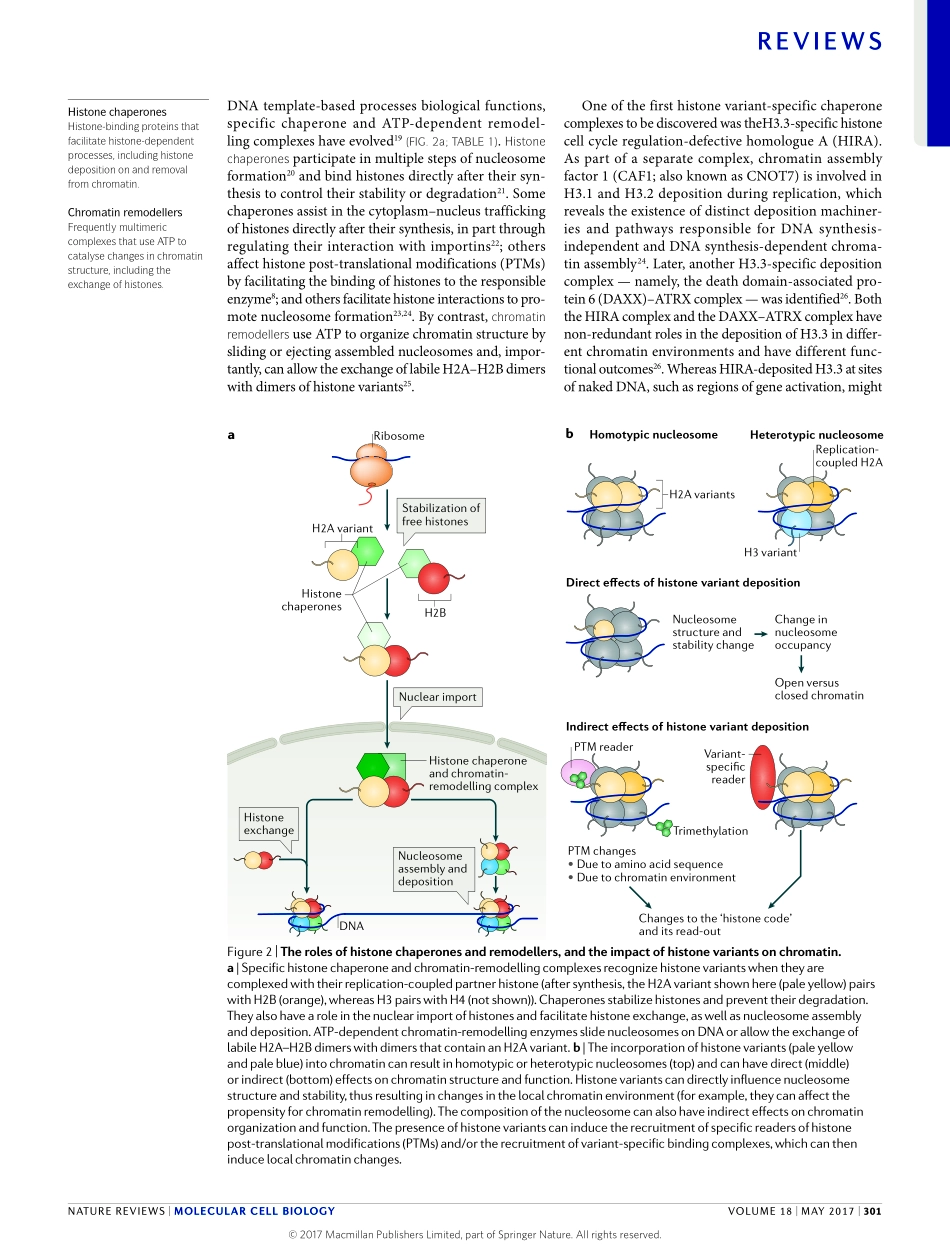
Oneofthemostabundantandmostconservedpro-teinfamiliesineukaryoticcellsisthehistonefamily.HistonespackagegeneticinformationintothenuclearspaceandcontributetotheregulationofallDNAtemplate-basedreactions.CorehistonesbindDNAaspartofthenucleosome,thebuildingblockofchro-matin,andlinkerhistonesbindtoDNAintheinter-nucleosomalspace.Asidefromtheso-called‘canonical’histones,whichcomprisethemajorityofanygivenhis-tonespeciesinanycell,evolutiondrovetheemergenceofhistonevariants,whichendowchromatinwithspecialpropertiesinalocus-specificmanner.Withrespecttothecorehistones,eightvariantsofH2A(H2A.X,H2A.Z.1,H2A.Z.2.1,H2A.Z.2.2,H2ABarrbodydefi-cient(H2A.Bbd;alsoknownasH2A.B),macroH2A1.1,macroH2A1.2andmacroH2A2)andsixvariantsofH3(H3.3,histoneH3-likecentromericproteinA(CENP-A),H3.1T,H3.5,H3.X(alsoknownasH3.Y.2)andH3.Y(alsoknownasH3.Y.1))havebeenidentifiedinhumansomaticcells.Inaddition,twotestis-specificvariantsofH2B(histoneH2BtypeWT(H2BFWT;alsoknownasH2B.W)andtestis-specifichistoneH2B(TSH2B;alsoknownashistoneH2Btype1A))havebeenfound.However,novariantsofH4haveyetbeendiscoveredinhighereukaryotes1.Surprisingly,inlowereukaryotessuchastrypanosomes2andsomeurochordates3,H4variantshavebeenfound,whichsug-gestseitherthatfunctionalspecializationofthishistonefamilyisevolutionarilypossibleorthatthesevariantsdoexistinhighereukaryotesbuttheirdetectionhassofarbeenunsuccessful.Notably,thedifferentvariantsaredistinctanduniqueintermsoftheirgeneandproteinsequences,aswellasthetimingoftheirtranscription,howtheirRNAisprocessedandwhentheyaredepos-itedonDNAduringthecellcycle(FIG.1).ThecanonicalhistonesaredepositedonDNAduringreplication,andinthisReviewwehereafterrefertotheseas‘replica-tion-coupled’histones.Eachofthereplication-coupledhistonesisencodedbymultiplegenesand,withafewexceptions,mostofthesegenesareorganizedintoclustersthroughoutthegenome4,5(FIG.1a).Thisprob-ablyensuresthattheexpressionofreplication-coupledhistones,whichtakesplaceduringS-phase,generateslargeandequalamountsofallfournucleosomecorehisto...