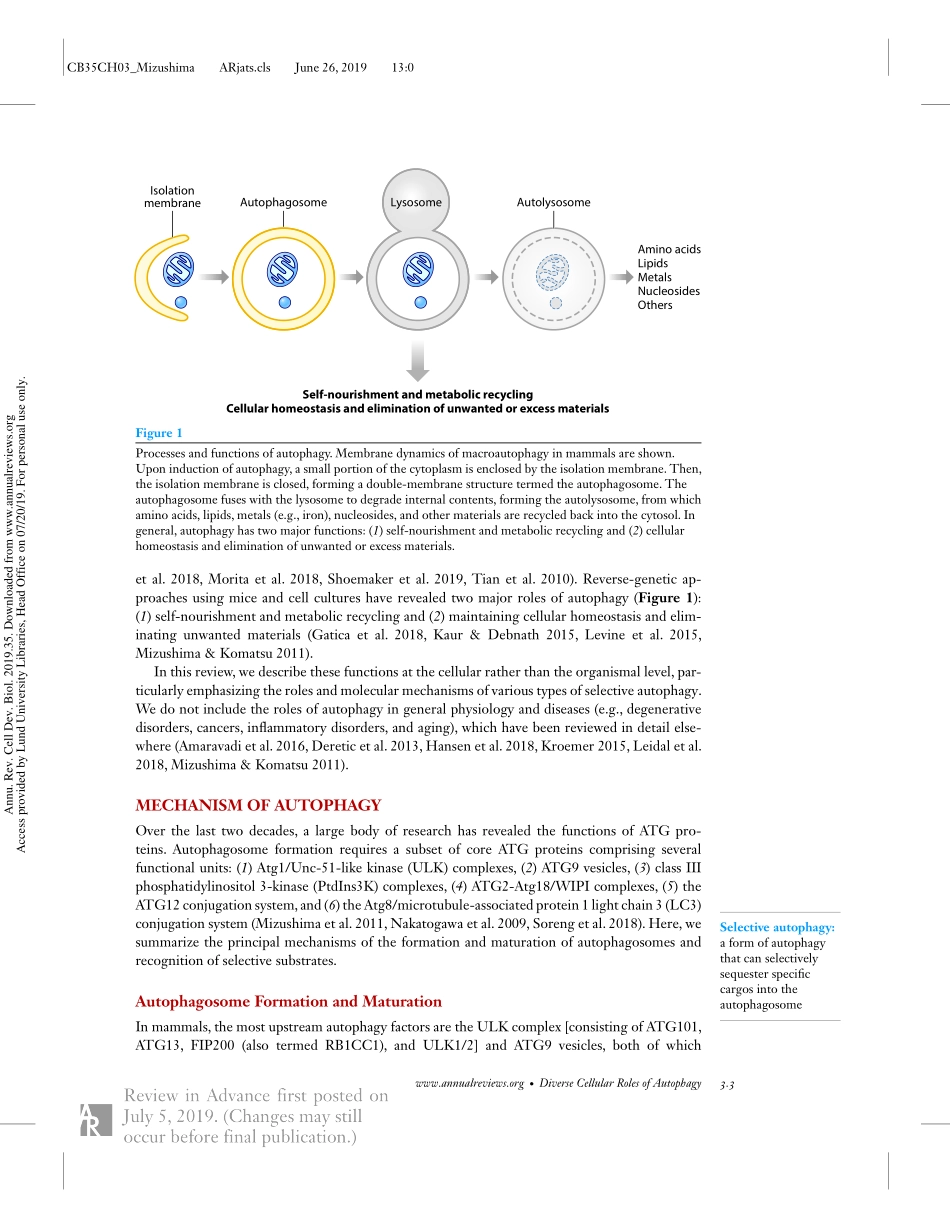
CB35CH03_MizushimaARjats.clsJune26,201913:0AnnualReviewofCellandDevelopmentalBiologyDiverseCellularRolesofAutophagyHideakiMorishita∗andNoboruMizushima∗DepartmentofBiochemistryandMolecularBiology,GraduateSchoolandFacultyofMedicine,TheUniversityofTokyo,Tokyo113-0033,Japan;email:morishita@m.u-tokyo.ac.jp,nmizu@m.u-tokyo.ac.jpAnnu.Rev.CellDev.Biol.2019.35:3.1–3.23TheAnnualReviewofCellandDevelopmentalBiologyisonlineatcellbio.annualreviews.orghttps://doi.org/10.1146/annurev-cellbio-100818-125300Copyright©2019byAnnualReviews.Allrightsreserved∗TheseauthorscontributedequallytothisarticleKeywordsautophagosome,selectiveautophagy,mitophagy,ER-phagy,nutrientstarvationAbstractMacroautophagyisanintracellulardegradationsystemthatdeliversdiversecytoplasmicmaterialstolysosomesviaautophagosomes.Recentadvanceshaveenabledidentificationofseveralselectiveautophagysubstratesandreceptors,greatlyexpandingourunderstandingofthecellularfunctionsofautophagy.Inthisreview,wedescribethediversecellularfunctionsofmacroautophagy,includingitsessentialcontributiontometabolicadapta-tionandcellularhomeostasis.Wealsodiscussemergingfindingsonthemechanismsandfunctionsofvarioustypesofselectiveautophagy.3.1ReviewinAdvancefirstpostedonJuly5,2019.(Changesmaystilloccurbeforefinalpublication.)Annu.Rev.CellDev.Biol.2019.35.Downloadedfromwww.annualreviews.orgAccessprovidedbyLundUniversityLibraries,HeadOfficeon07/20/19.Forpersonaluseonly.CB35CH03_MizushimaARjats.clsJune26,201913:0Autophagosome:adouble-membranestructureenclosingcytoplasmicmaterialsContentsINTRODUCTION..............................................................3.2MECHANISMOFAUTOPHAGY................................................3.3AutophagosomeFormationandMaturation......................................3.3SelectiveAutophagy............................................................3.4APPROACHESTOUNDERSTANDINGCELLULARROLESOFAUTOPHAGY.............................................................3.6MonitoringofAutophagy...........................