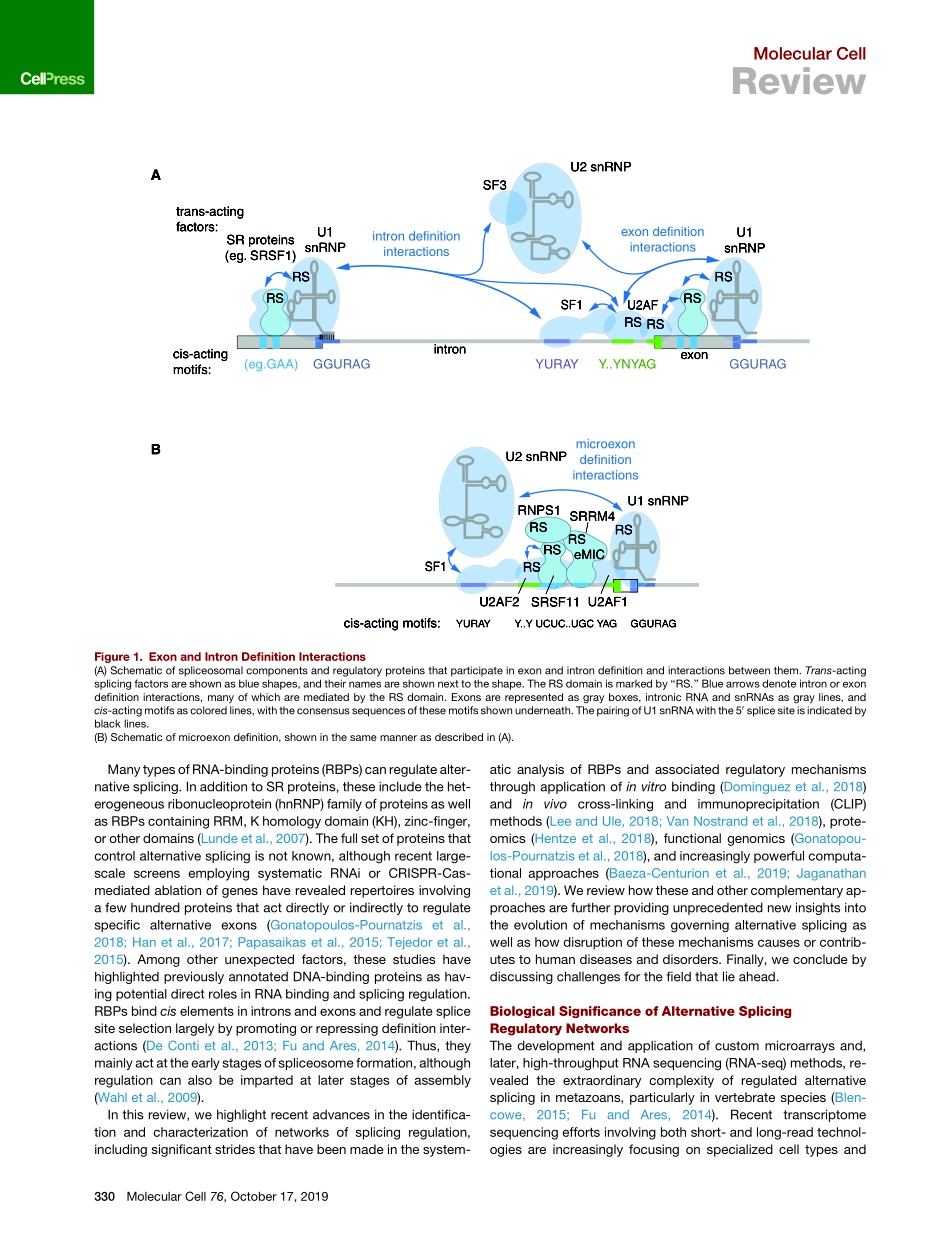
MolecularCellReviewAlternativeSplicingRegulatoryNetworks:Functions,Mechanisms,andEvolutionJernejUle1,2,*andBenjaminJ.Blencowe3,4,*1TheFrancisCrickInstitute,LondonNW11AT,UK2DepartmentofNeuromuscularDiseases,UCLQueenSquareInstituteofNeurology,QueenSquare,LondonWC1N3BG,UK3DonnellyCentre,UniversityofToronto,Toronto,ONM5S3E1,Canada4DepartmentofMolecularGenetics,UniversityofToronto,Toronto,ON,M5S3E1,Canada*Correspondence:jernej.ule@crick.ac.uk(J.U.),b.blencowe@utoronto.ca(B.J.B.)https://doi.org/10.1016/j.molcel.2019.09.017High-throughputsequencing-basedmethodsandtheirapplicationsinthestudyoftranscriptomeshaverevolutionizedourunderstandingofalternativesplicing.Networksoffunctionallycoordinatedandbiologi-callyimportantalternativesplicingeventscontinuetobediscoveredinanever-increasingdiversityofcelltypesinthecontextofphysiologicallynormalanddiseasestates.Thesestudieshavebeencomplementedbyeffortsdirectedatdefiningsequencecodesgoverningsplicingandtheircognatetrans-actingfactors,whichhaveilluminatedimportantcombinatorialprinciplesofregulation.Additionalstudieshaverevealedcriticalrolesofposition-dependent,multivalentprotein-RNAinteractionsthatdirectsplicingoutcomes.In-vestigationsofevolutionarychangesinRNAbindingproteins,splicevariants,andassociatedciselementshavefurthershedlightontheemergence,mechanisms,andfunctionsofsplicingnetworks.Progressintheseareashasemphasizedtheneedforacoordinated,community-basedefforttosystematicallyaddressthefunctionsofindividualsplicevariantsassociatedwithnormalanddiseasebiology.Transcriptsfromnearlyallhumanprotein-codinggenesundergooneormoreformsofalternativesplicing,suchasinclusionorskippingofindividual‘‘cassette’’exons,switchingbetweenalternative50and30splicesites,differentialretentionofintrons,mutuallyexclusivesplicingofadjacentexons,andother,morecomplexpatternsofsplicesiteselection(Panetal.,2008;Wangetal.,2008).Alloftheseformsofsplicingrequirethespliceosome,amegadaltonmachinethatcatalyzessplicingre-actions(Wahletal.,2009).Spliceosomeforma...