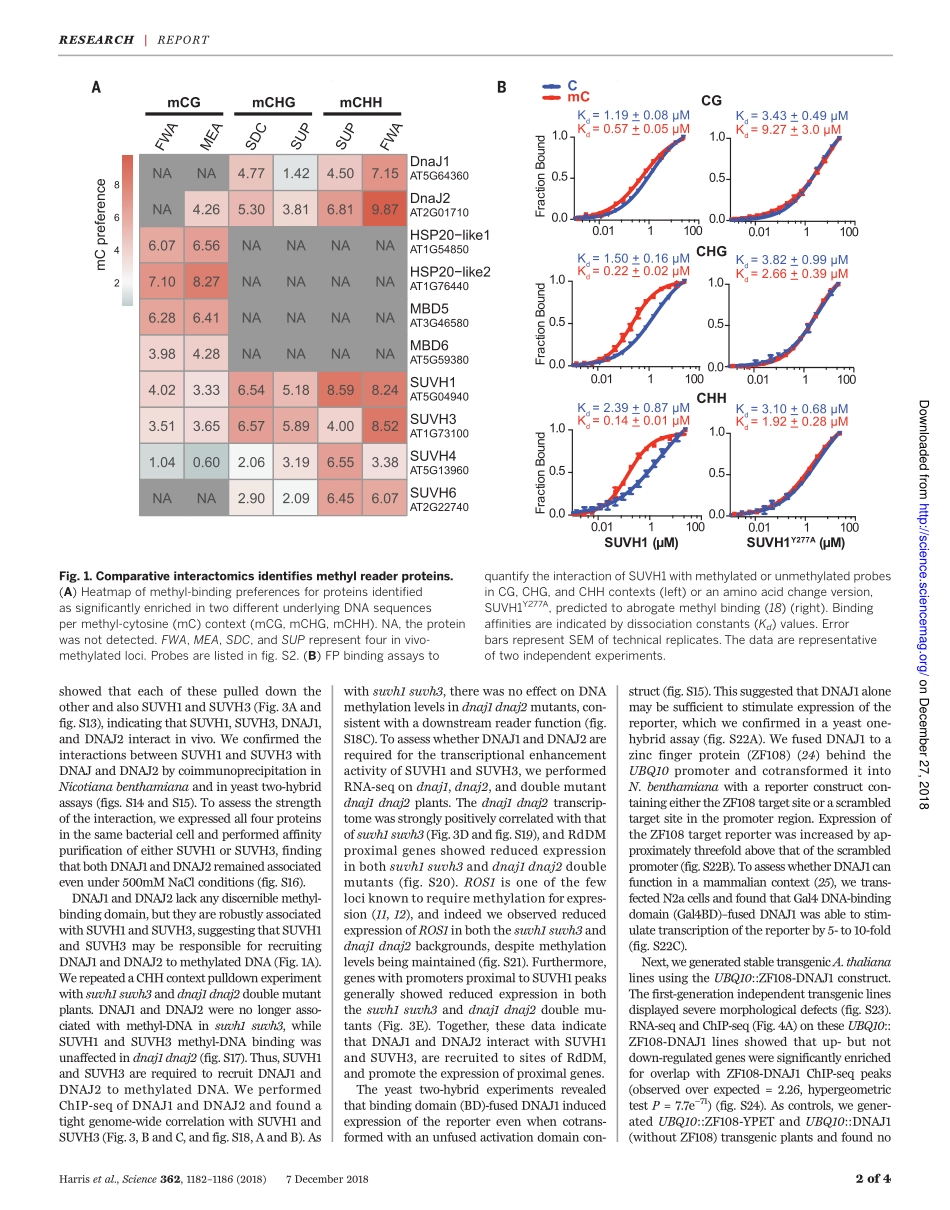
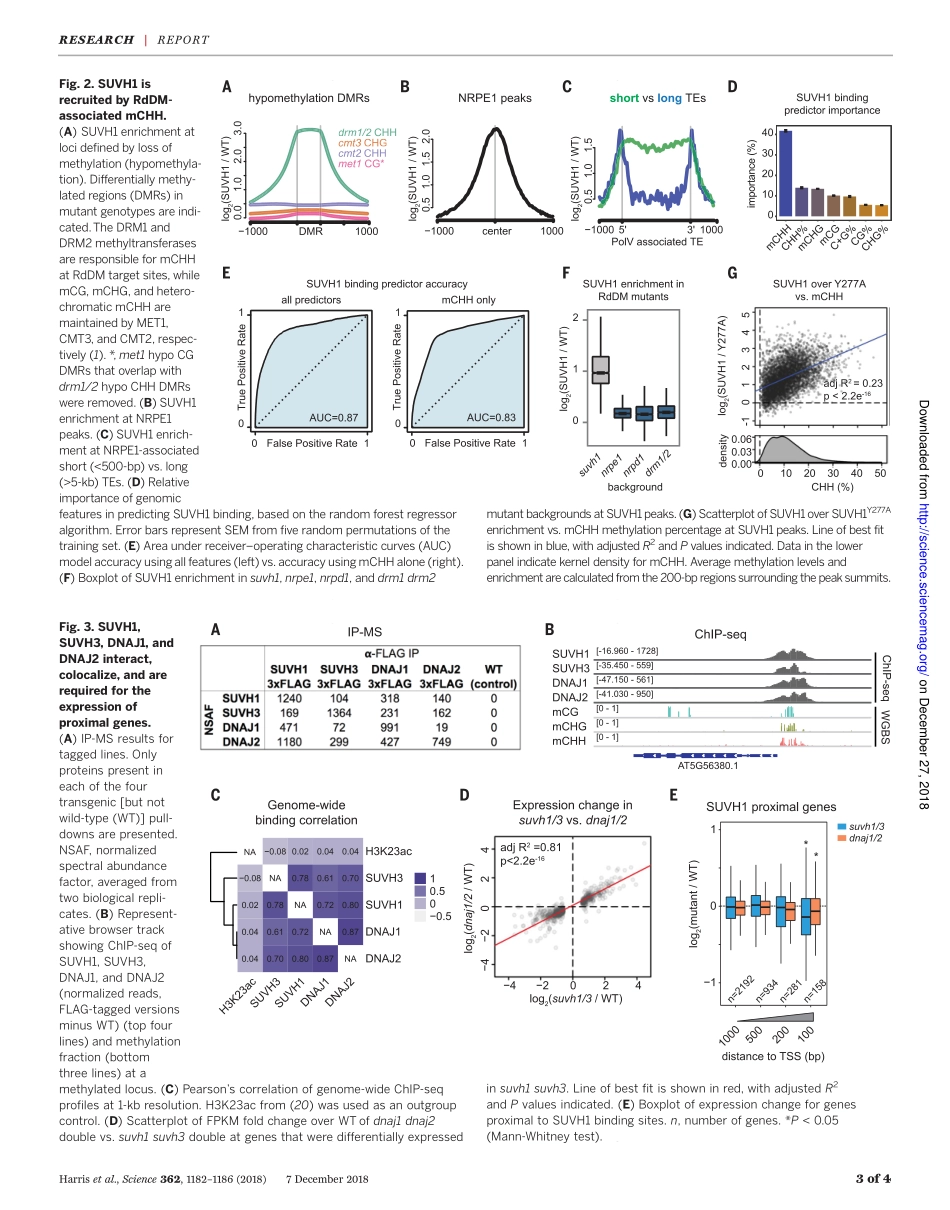
MOLECULARBIOLOGYADNAmethylationreadercomplexthatenhancesgenetranscriptionC.JakeHarris1*,MarionScheibe2*,SomsakulPopWongpalee1†,WanluLiu1,EvanM.Cornett3,RobertM.Vaughan3,XueqinLi4,WeiChen4,YanXue1,ZhenhuiZhong1,5,LindaYen1,WilliamD.Barshop6,ShimaRayatpisheh6‡,JavierGallego-Bartolome1,MartinGroth1§,ZonghuaWang5,7,JamesA.Wohlschlegel6,JiamuDu4,ScottB.Rothbart3,FalkButter2||,StevenE.Jacobsen1,8||DNAmethylationgenerallyfunctionsasarepressivetranscriptionalsignal,butitisalsoknowntoactivategeneexpression.Ineithercase,thedownstreamfactorsremainlargelyunknown.Byusingcomparativeinteractomics,weisolatedproteinsinArabidopsisthalianathatassociatewithmethylatedDNA.TwoSU(VAR)3-9homologs,thetranscriptionalantisilencingfactorSUVH1,andSUVH3,wereamongthemethylreadercandidates.SUVH1andSUVH3boundmethylatedDNAinvitro,wereassociatedwitheuchromaticmethylationinvivo,andformedacomplexwithtwoDNAJdomain-containinghomologs,DNAJ1andDNAJ2.EctopicrecruitmentofDNAJ1enhancedgenetranscriptioninplants,yeast,andmammals.Thus,theSUVHproteinsbindtomethylatedDNAandrecruittheDNAJproteinstoenhanceproximalgeneexpression,therebycounteractingtherepressiveeffectsoftransposoninsertionneargenes.DNAmethylationfrequentlymarkstrans-posableelements(TEs)ineukaryoticge-nomes(1–3).Inplants,theRNA-directedDNAmethylation(RdDM)pathwayisre-sponsiblefortheinitialestablishmentofmethylationinCG,CHG,andCHHcontexts(4).TEinsertionscanexertatranscriptionaleffectonneighboringgenes(5–8),andpromotermeth-ylationistypicallyassociatedwithgenerepres-sion(9).However,exceptionsexistwherepromotermethylationisrequiredforgeneexpression(10–14).Thedownstreamfactorsthatperceivemeth-ylationtomediatethesedivergenttranscrip-tionaleffectsarestillpoorlycharacterized,andlittleisknownofhowmethylationcanstimu-lategenetranscription.ToidentifyproteinsinArabidopsisthalianathatrecognizemethylatedDNA,weincubatednuclearextractfromfloralbudtissuewitheithermethylatedorunmethylatedbiotinylateddouble-strandedDNAoligonucleotides,affinitypurifiedtheDNA,ands...