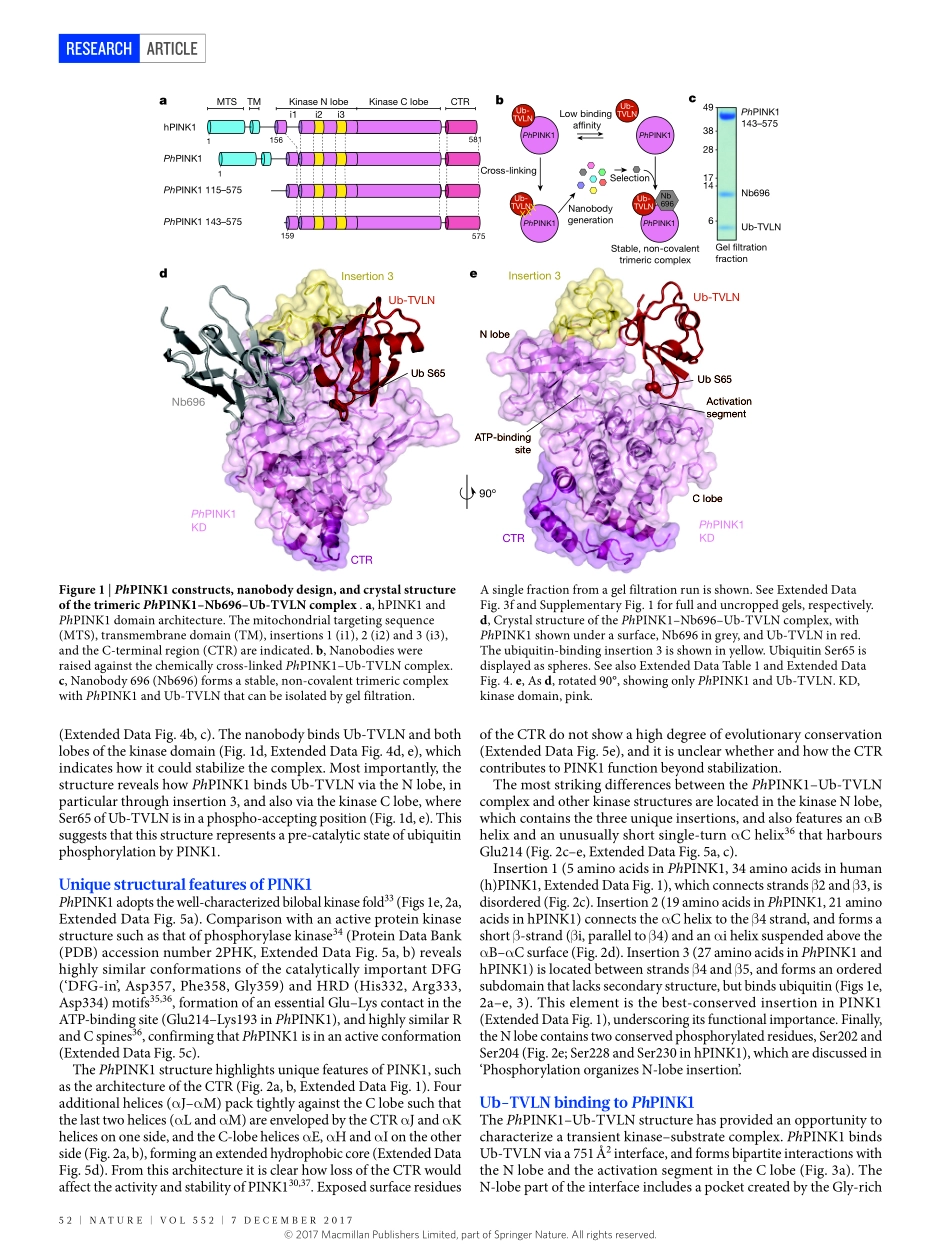
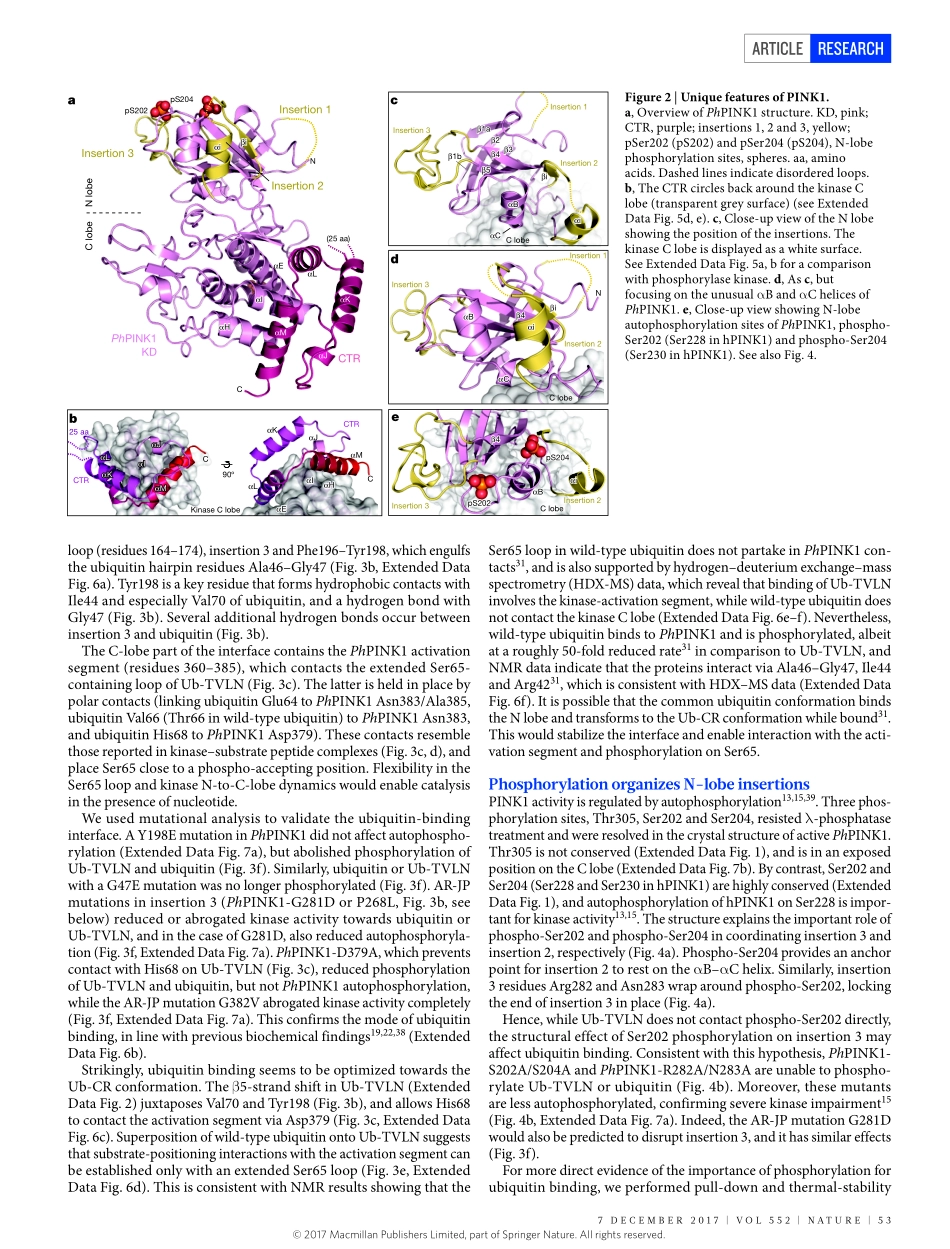
7DECEMBER2017|VOL552|NATURE|51ARTiCLEdoi:10.1038/nature24645StructureofPINK1incomplexwithitssubstrateubiquitinAlexanderF.Schubert1,ChristinaGladkova1,ElsPardon2,3,JaneL.Wagstaff1,StefanM.V.Freund1,JanSteyaert2,3,SarahL.Maslen1&DavidKomander1Parkinson’sdisease(PD)isaneurodegenerativeconditioncausedbythelossofdopaminergicneuronsinthesubstantianigrathatmanifestsclinicallyintheformofcharacteristicmotordefects1.MostcasesofPDaresporadicandfoundinpeopleabovetheageof60,butroughly10%ofcasesarehereditaryformswithanearlieronset.AutosomalrecessivejuvenileParkinsonism(ARJP)iscausedbymutationsinaround15PARKgenesthathavebeenannotatedtodate2.TheSer/ThrkinasePINK1wasoneofthefirstPARKgenestobeidentified3.Mutationofpink1inDrosophilacausedPDlikesymptoms,andlinkedPINK1totheE3ubiquitinligaseParkin4–8.DetailedfunctionalreconstructionhassincelinkedPINK1andParkintomitophagy,animportantmitochondrialqualitycontrolprocess9–11.MitochondrialdamageandsubsequentlossofmitochondrialmembranepotentialleadstothestabilizationofPINK1onthemitochondrialoutermembrane(MOM)12andsubsequentdimerization,autophosphorylationandactivationofPINK113–16.PINK1activityleadstophosphorylationofubiquitinonSer65(hereafterreferredtoasphosphoubiquitin)17–21,andtophosphorylationoftheUbldomainofParkinonitsstructurallyidenticalSer65residue16,21–25.Together,phosphorylationandphosphoubiquitinbindingleadtolocalizedParkinactivationonmitochondria,whereParkinubiquitinatesMOMproteins26,triggeringafeedforwardmechanism21.Mitochondrialubiquitinationrecruitsmitophagyadaptorsandinitiatesclearanceofthedamagedmitochondria9,10,27,28.PINK1isoneofthemostdivergenthumanproteinkinases29.ItcontainsanNterminalmitochondriatargetingandtransmembranesegment,anunconservedregion,akinasedomainwiththreeinsertionsintheNlobe,andaconservedCterminalregion(CTR)ofunknownfunctionandstructure(Fig.1a,ExtendedDataFig.1).Althoughdistinctfromotherkinases,PINK1ishighlyconservedacrossspecies(ExtendedDataFig.1),whichhasledtotheidentificat...