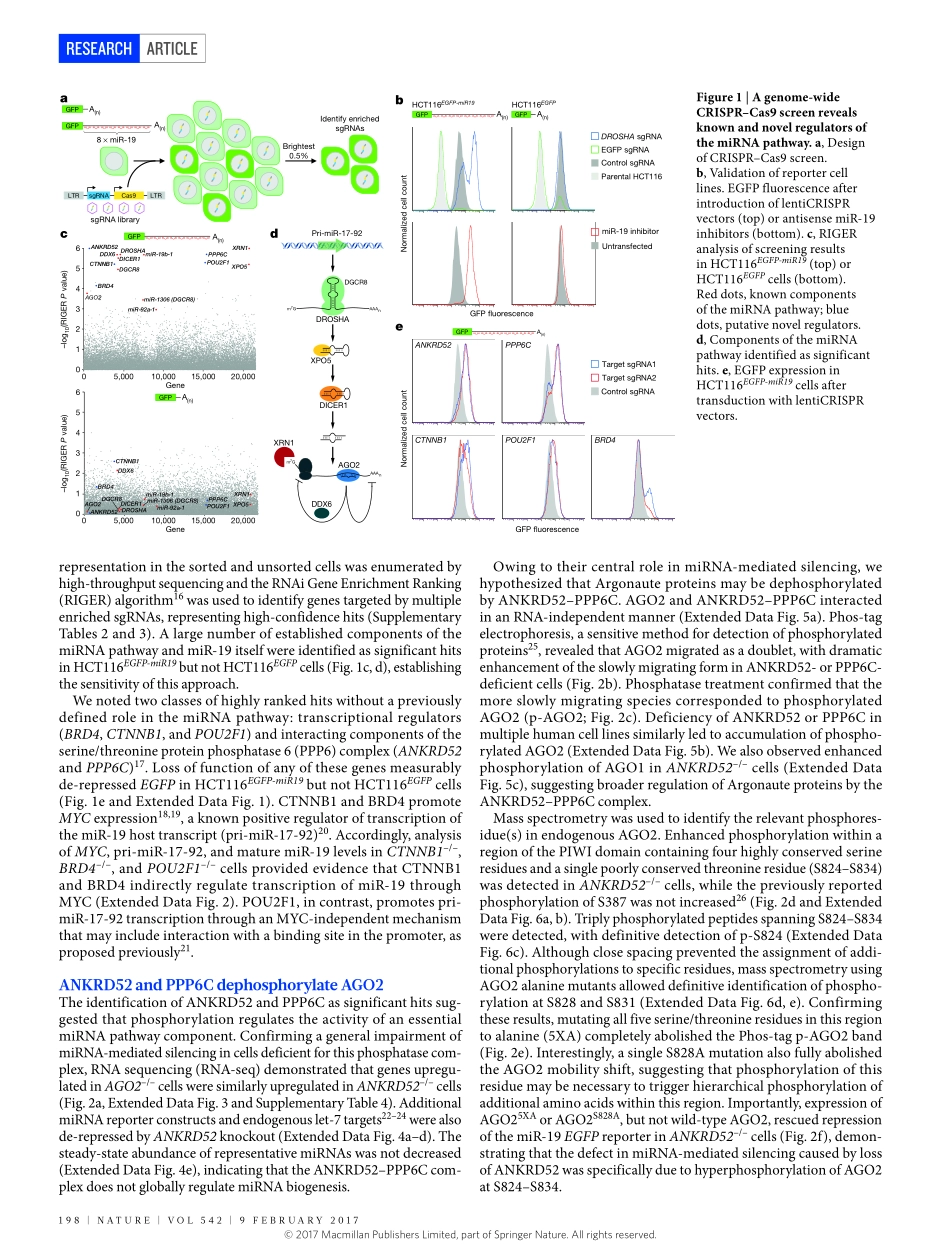
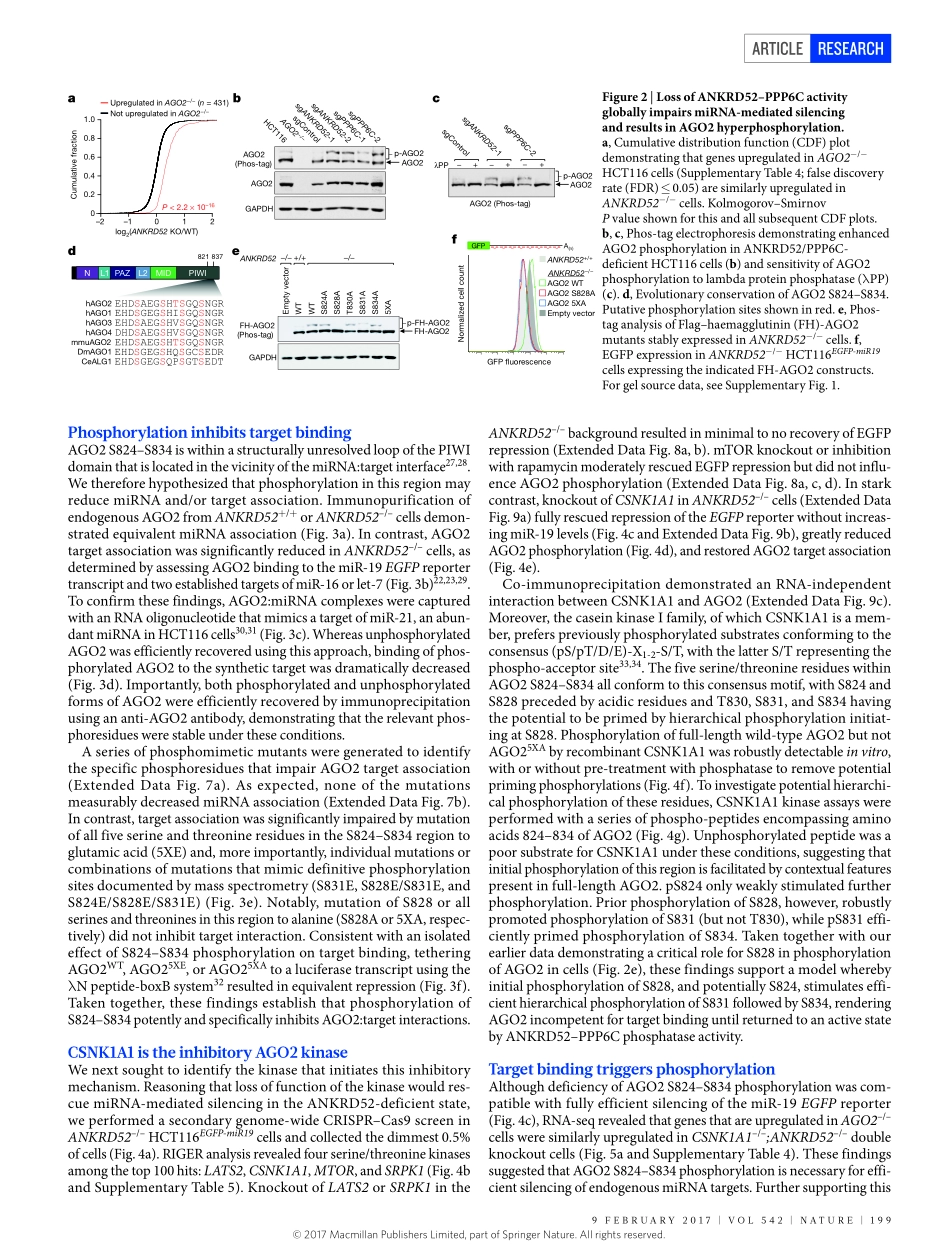
9february2017|VOL542|NaTure|197arTicLedoi:10.1038/nature21025AnArgonautephosphorylationcyclepromotesmicroRNA-mediatedsilencingryanJ.Golden1,2,beibeichen3,4,TuoLi1,Julianebraun1,HemaManjunath1,Xiangchen1,JiaxiWu5,VanessaSchmid6,Tsung-chengchang1,florianKopp1,andresramirez-Martinez1,VincentS.Tagliabracci1,ZhijianJ.chen1,7,yangXie3,4,8&JoshuaT.Mendell1,7,8,9ThemiRNApathwayisessentialfordevelopmentandhomeostasisindiversespecies1,2.miRNAsassociatewithArgonaute(AGO)proteins,whichtheyguidetopartiallycomplementarysitesinmessengerRNAs(mRNAs)3,leadingtoreducedstabilityandtranslationoftargetedmessages4.miRNAsselecttargetsprimarilythroughbasepairingoftheirseedregions,nucleotides2–7.Consequently,thepotentialtargetrepertoireforagivenmiRNAisvast.Multiplesequencefeaturesofbonafidetargetsitesdistinguishthemfromnon-functionalsiteswithseedcomplementarity5.Nevertheless,recentexperimentshavedemon-stratedthatthefunctionalpooloftargetsgreatlyexceedsthequantityofmiRNAsinmammaliancells6,7.Whiletheintrinsicsequencechar-acteristicsofmiRNAbindingsitesstronglyinfluencethekineticsofAGO:targetinteractions8,9,itiscurrentlyunknownwhetheradditional,activemechanismsexistthatinfluencemRNAbindingtofacilitatenav-igationoftheextensivetargetlandscape.RNAinterference(RNAi)screenshavebeenusedtodissectthemiRNApathwayininvertebrates10,11.Analogousexperimentsinhumancells,however,havebeenhinderedbythefactthattheRNAiandmiRNApathwayssharecommonmolecularmachinery.Thislim-itationmaybecircumventedbyrecentadvancesinCRISPR-mediatedgenomeediting,whichoffersarobustalternativeforgeneticloss-of-functionscreensinhumancells12,13.HerewedescribetheapplicationofCRISPR–Cas9screeningtoidentifynovelregulatorsofmiRNA-me-diatedsilencing.TheseexperimentsrevealthattheANKRD52–PPP6CphosphatasecomplexperformsacriticalfunctioninthemiRNApath-waybydephosphorylatingasetofhighlyconservedaminoacidsinAGO2.Asecondarygenome-widescreenrevealedCSNK1A1asthekinasethatphosphorylatesAGO2onthesesites.ThisAGO2phospho-rylationcycleistriggeredbytarg...