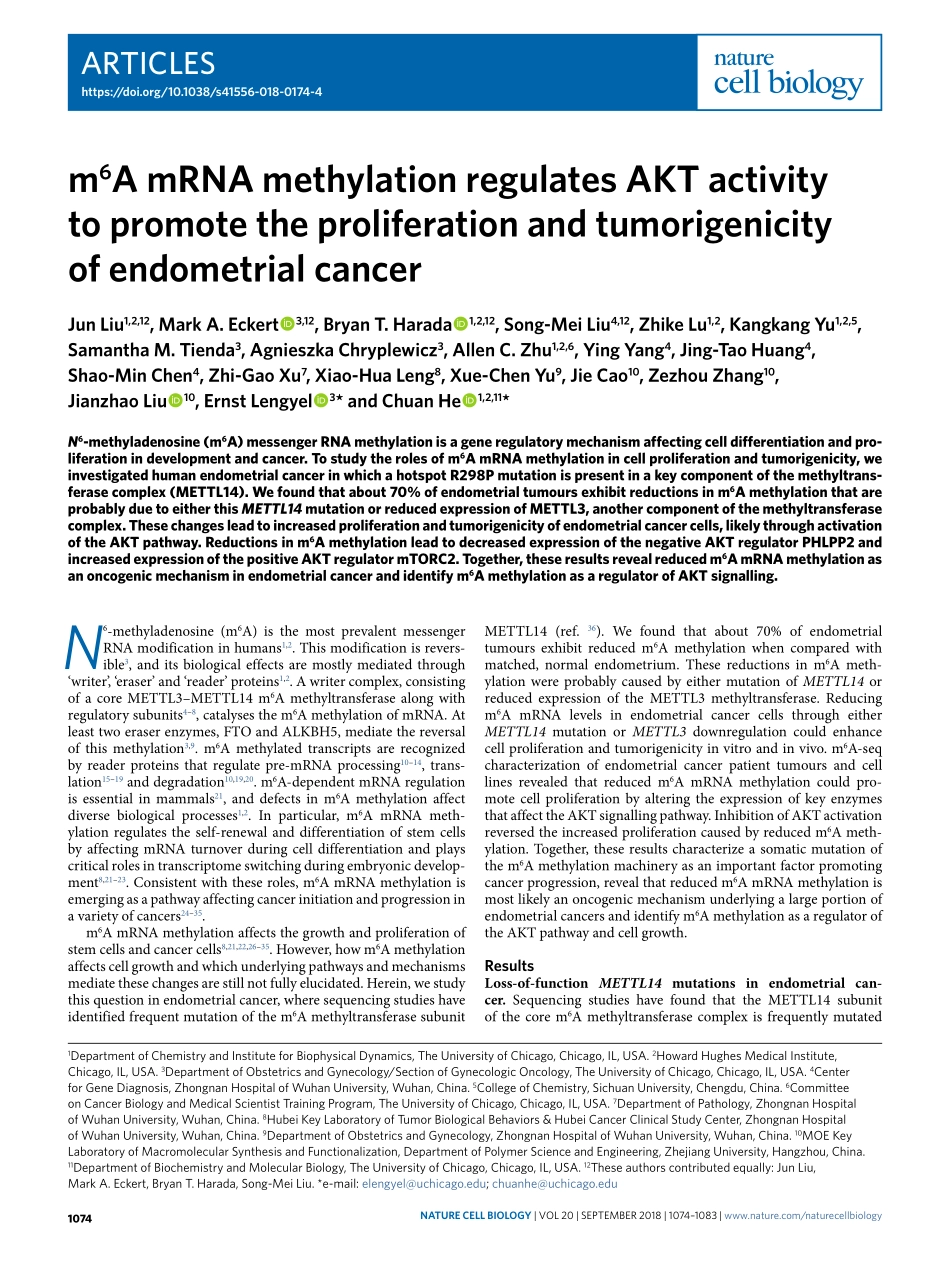
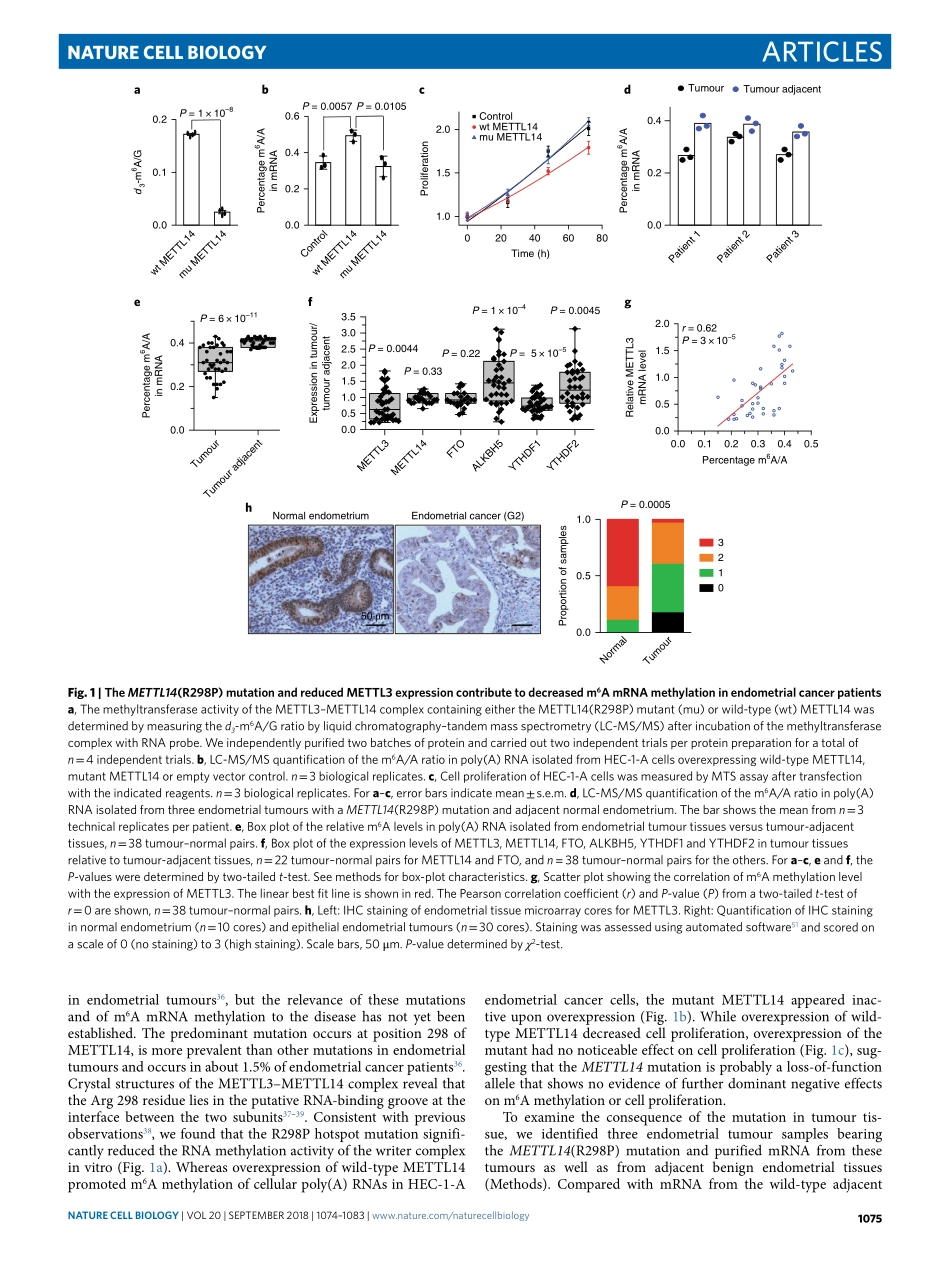
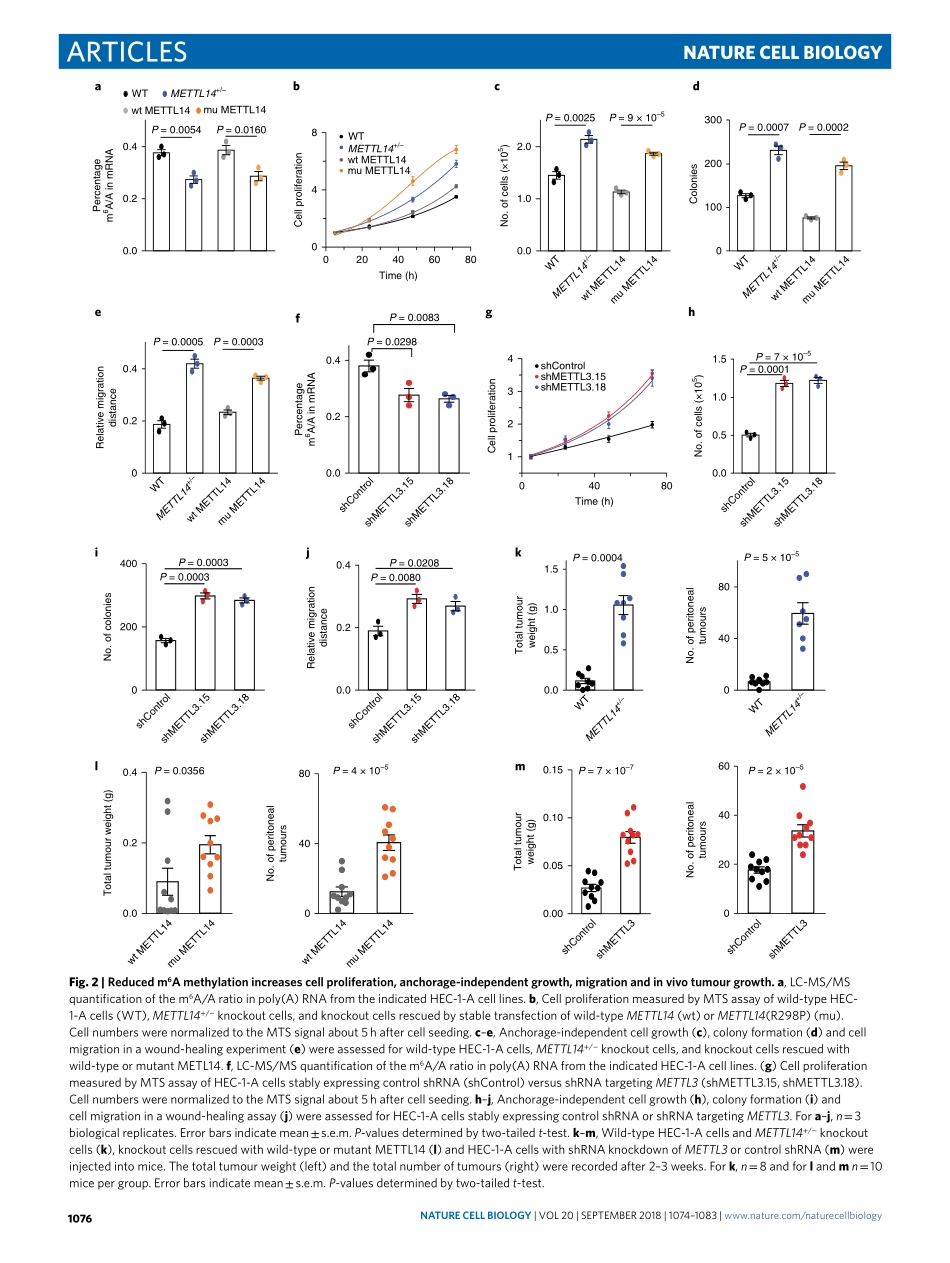
Articleshttps://doi.org/10.1038/s41556-018-0174-41DepartmentofChemistryandInstituteforBiophysicalDynamics,TheUniversityofChicago,Chicago,IL,USA.2HowardHughesMedicalInstitute,Chicago,IL,USA.3DepartmentofObstetricsandGynecology/SectionofGynecologicOncology,TheUniversityofChicago,Chicago,IL,USA.4CenterforGeneDiagnosis,ZhongnanHospitalofWuhanUniversity,Wuhan,China.5CollegeofChemistry,SichuanUniversity,Chengdu,China.6CommitteeonCancerBiologyandMedicalScientistTrainingProgram,TheUniversityofChicago,Chicago,IL,USA.7DepartmentofPathology,ZhongnanHospitalofWuhanUniversity,Wuhan,China.8HubeiKeyLaboratoryofTumorBiologicalBehaviors&HubeiCancerClinicalStudyCenter,ZhongnanHospitalofWuhanUniversity,Wuhan,China.9DepartmentofObstetricsandGynecology,ZhongnanHospitalofWuhanUniversity,Wuhan,China.10MOEKeyLaboratoryofMacromolecularSynthesisandFunctionalization,DepartmentofPolymerScienceandEngineering,ZhejiangUniversity,Hangzhou,China.11DepartmentofBiochemistryandMolecularBiology,TheUniversityofChicago,Chicago,IL,USA.12Theseauthorscontributedequally:JunLiu,MarkA.Eckert,BryanT.Harada,Song-MeiLiu.*e-mail:elengyel@uchicago.edu;chuanhe@uchicago.eduN6-methyladenosine(m6A)isthemostprevalentmessengerRNAmodificationinhumans1,2.Thismodificationisrevers-ible3,anditsbiologicaleffectsaremostlymediatedthrough‘writer’,‘eraser’and‘reader’proteins1,2.Awritercomplex,consistingofacoreMETTL3–METTL14m6Amethyltransferasealongwithregulatorysubunits4–8,catalysesthem6AmethylationofmRNA.Atleasttwoeraserenzymes,FTOandALKBH5,mediatethereversalofthismethylation3,9.m6Amethylatedtranscriptsarerecognizedbyreaderproteinsthatregulatepre-mRNAprocessing10–14,trans-lation15–19anddegradation10,19,20.m6A-dependentmRNAregulationisessentialinmammals21,anddefectsinm6Amethylationaffectdiversebiologicalprocesses1,2.Inparticular,m6AmRNAmeth-ylationregulatestheself-renewalanddifferentiationofstemcellsbyaffectingmRNAturnoverduringcelldifferentiationandplayscriticalrolesintranscriptomeswitchingduringembryonicdevelop-ment...