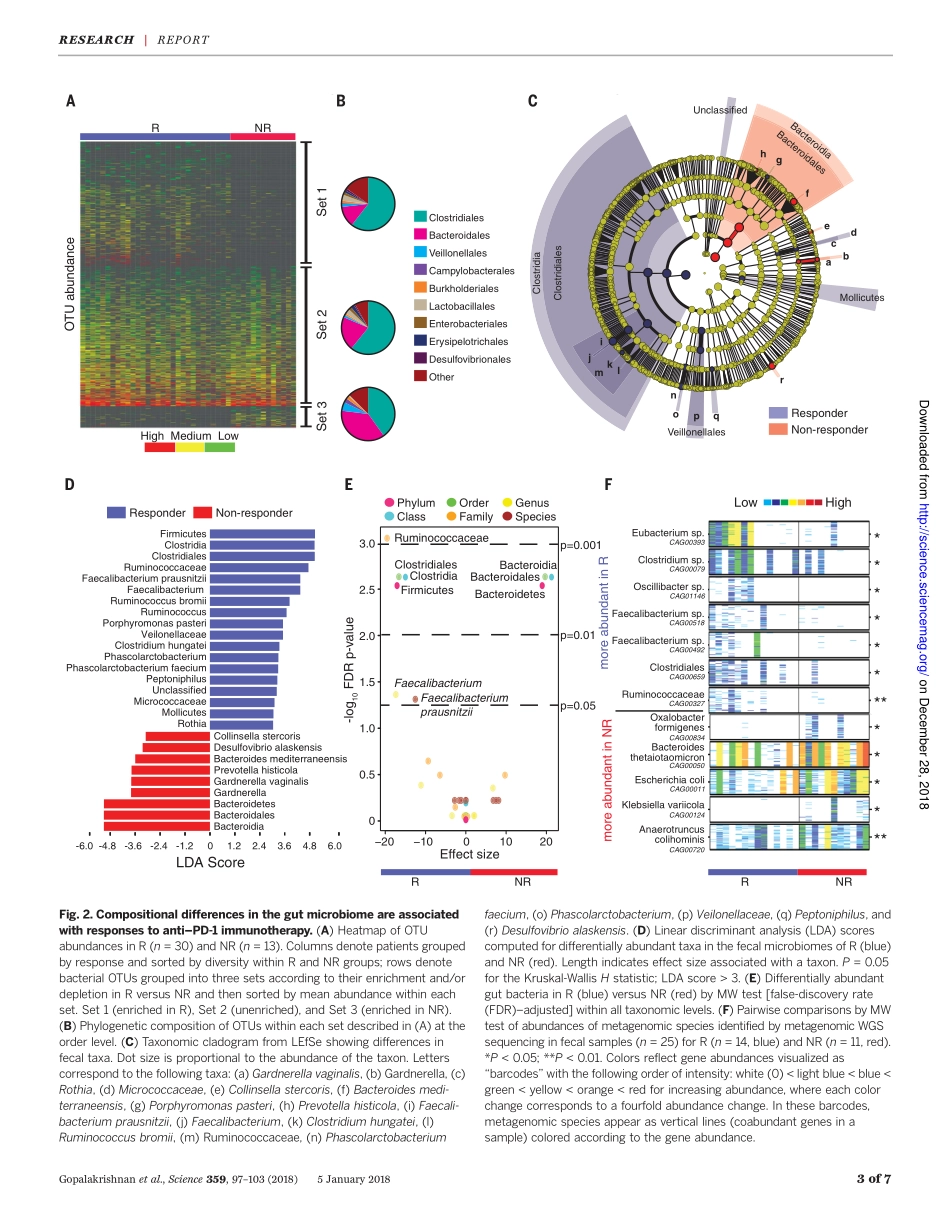
CANCERIMMUNOTHERAPYGutmicrobiomemodulatesresponsetoanti–PD-1immunotherapyinmelanomapatientsV.Gopalakrishnan,1,2*C.N.Spencer,2,3*L.Nezi,3*A.Reuben,1M.C.Andrews,1T.V.Karpinets,3P.A.Prieto,1†D.Vicente,1K.Hoffman,4S.C.Wei,5A.P.Cogdill,1,5L.Zhao,3C.W.Hudgens,6D.S.Hutchinson,7T.Manzo,3M.PetacciadeMacedo,6‡T.Cotechini,8T.Kumar,3W.S.Chen,9S.M.Reddy,10R.SzczepaniakSloane,1J.Galloway-Pena,11H.Jiang,1P.L.Chen,9§E.J.Shpall,12K.Rezvani,12A.M.Alousi,12R.F.Chemaly,11S.Shelburne,3,11L.M.Vence,5P.C.Okhuysen,11V.B.Jensen,13A.G.Swennes,7F.McAllister,14E.MarceloRiquelmeSanchez,14Y.Zhang,14E.LeChatelier,15L.Zitvogel,16N.Pons,15J.L.Austin-Breneman,1||L.E.Haydu,1E.M.Burton,1J.M.Gardner,1E.Sirmans,17J.Hu,18A.J.Lazar,6,9T.Tsujikawa,8A.Diab,17H.Tawbi,17I.C.Glitza,17W.J.Hwu,17S.P.Patel,17S.E.Woodman,17R.N.Amaria,17M.A.Davies,17J.E.Gershenwald,1P.Hwu,17J.E.Lee,1J.Zhang,3L.M.Coussens,8Z.A.Cooper,1,3¶P.A.Futreal,3C.R.Daniel,4,2N.J.Ajami,7J.F.Petrosino,7M.T.Tetzlaff,6,9P.Sharma,5,19J.P.Allison,5R.R.Jenq,3#J.A.Wargo1,3#**Preclinicalmousemodelssuggestthatthegutmicrobiomemodulatestumorresponsetocheckpointblockadeimmunotherapy;however,thishasnotbeenwell-characterizedinhumancancerpatients.Hereweexaminedtheoralandgutmicrobiomeofmelanomapatientsundergoinganti–programmedcelldeath1protein(PD-1)immunotherapy(n=112).Significantdifferenceswereobservedinthediversityandcompositionofthepatientgutmicrobiomeofrespondersversusnonresponders.Analysisofpatientfecalmicrobiomesamples(n=43,30responders,13nonresponders)showedsignificantlyhigheralphadiversity(P<0.01)andrelativeabundanceofbacteriaoftheRuminococcaceaefamily(P<0.01)inrespondingpatients.Metagenomicstudiesrevealedfunctionaldifferencesingutbacteriainresponders,includingenrichmentofanabolicpathways.Immuneprofilingsuggestedenhancedsystemicandantitumorimmunityinrespondingpatientswithafavorablegutmicrobiomeaswellasingerm-freemicereceivingfecaltransplantsfromrespondingpatients.Together,thesedatahaveimportantimplicationsforthetreatmentofmelanomapatientswithi...